Abstract
Image voxel/vertex-wise feature in the brain is widely used for automatic classification or significant region detection of various dementia syndromes. In these studies, the non-imaging variables, such as age, will affect the results, but may be uninterested to the clinical applications. Imaging data can be considered as a combination of the confound variable (e.g. age) and the variable of clinical interest (e.g. AD diagnosis). However, non-imaging confound variable is not well dealt in each voxel. In this paper, we proposed a spatial-anatomical regularized parametric function fitting approach that explicitly modeling the relationship between the voxelwise feature and the confound variable. By adding the spatial-anatomical regularization, our model not only obtains a better voxelwise feature estimation, but also generates a more interpretable parameter map to help understand the effect of confound variable on imaging features. Besides the commonly used linear model, we also develop a spatial-anatomical regularized voxelwise general logistic model to investigate deeper of the aging process in gray matter and white matter density map.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Neuroimaging data (e.g. MRI) is widely used to predict clinical or psychometric interest variable (MMSE sore or AD diagnosis) [1, 2]. It is believed that brain pathology or psychology change will lead to brain morphology [3] or functional [3] change in the neuroimaging data. However, these changes can be considered as combinations of effect of the clinical interested variable (e.g. Alzheimer’s Disease) and other factors (e.g. age). Such variables are commonly referred as “Confound" variables, which are highly correlated with the neuroimaging data but independent from the clinical interested variable. Such confound variables may decrease the power of the prediction or significant region detection [4].
To remove the effect of the confound variables, many approaches have been proposed to learn a better predictor. The most common approach is to match the sample subject with respect to the confound variable. However, such approach will largely decrease the number of variable in imaging data and limit the usage of the learned predictor. Another approach is to combine the confound variables and the features from image to jointly learn a predictor [5, 6]. However, in many methods, especially for methods using voxelwise feature, the number of imaging feature is much larger than the size of confound variables. This unbalance will likely decrease the correction effect of the confound variables. Lastly, the influence of the confound variables can be filtered out by independently fitting a linear model of the confound variables using the normal control subject and remove them from the image features [7]. After filtering out the image features related to the confound variables, the predictor can focus on the features related to the clinical interested variable and will be not sensitive to the length of features.
Inspired by the success of the filtering out approach [7], we proposed a spatial-anatomical regularized voxelwise model fitting approach to model the effect of the confound variables to the imaging feature. Besides the better estimation of confound variables related feature, the proposed method improves the interpretability of the learned voxelwise model. Another contribution of the proposed method is the portability to different models. Compared to the voxelwise independent linear model used in [7], our approach obtains more influence of the confound variables. In our application, we show the relationship between the voxelwise gray/white density and the age in normal subjects.
2 Methods
In this section, we first define the notations before explaining the details. The \(F_i(x)\) represents the voxelwise feature in i-th sample subject at voxel x, \(i \in \{1,2,\cdots ,N\}\). The \(\varvec{\theta }_k(x)\) represents the value of the k-th parameter map at voxel x, \(k \in \{1,2,\cdots ,K\}\). The K is the number of parameters for each voxel, and it varies for different models, e.g. \(K=2\) is for the linear model while \(K=4\) is for the general logistic model. The \(t_i\) represents the age of a sample subject i when the scan was taken.
2.1 Age Correction in Voxel Level
For each voxel x, we assume that the feature \(F_i(x)\) is the combination of the clinical interested variable and the confound variables. In our application, we consider \(F_i(x)=D_i(x)+A(x,t_i)\), where \(D_i(x)\) is the voxelwise feature related to the disease progress and \(A(x,t_i)\) the voxelwise feature related to the normal aging progress. For a normal subject, we can assume that there is no disease related progress and \(F_i(x)=0+A(x,t_i)\). Therefore, the age correction problem is simplified to model the normal aging part \(A(x,t_i)\) by a parametric function, with parameter \(\varvec{\theta }\).
Due to the large variations of the brain changes, it is reasonable to assume that different brain regions have different parameters. A voxelwise generative model \(f(t_i; \varvec{\theta }(x))\) is built to estimate the normal aging effect and the estimation error over all normal sample subjects observation \(F_i(x)\) is minimized. The optimal parameter of a linear model is estimated using:
The linear function is too weak to model the complex brain changes of normal aging, so a general logistic function with 4 parameters is introduced to model the normal aging related feature. In the general logistic function:
the parameter \(T_0\) is the time point where the curve change has the highest speed and it is often considered as the key time point to indicate the starting time. The parameter \(\alpha \) represents the growth rate of the curve. A large \(\alpha \) indicates that the age related change appears in a very short time interval. The M and N limit the range of the response of f(t). Therefore, even for a new sample with age t far from the training set, the general logistic is opt to generate a reasonable response, while the linear model may generate an unreasonable higher or lower response. For the general logistic function, the optimal parameter \(\varvec{\theta }(x)\) is estimated using:
2.2 Spatial-Anatomical Regularization
The voxelwise estimation of the optimal parameter commonly ignores the spatial and anatomical relation among voxels, however, these relations are very important in many medical image analysis applications. For example, the parameters from the voxelwise function fitting can be used to segment the myocardial scar identification [8] and the integration of the spatial-anatomical relations among voxels can improve the classification performance [9]. A spatial-anatomical regularization (SAR) penalty term was integrated in our proposed method which encourages the smoothness of the parameters from spatial-anatomical neighbor voxels.
Given an anatomical segmentation \(\varvec{S}\), the spatial-anatomical neighbor voxel relation R(p, q) between point pair \(\{p,q\}\) is defined as:
and the SAR penalty is computed as:
The SAR penalty can be simplified as a quadratic term:
where \(\varvec{L}\) is a sparse matrix. Each row of \(\varvec{L}\) encodes one pair of spatial-anatomical neighbor point pair. For the c-th spatial-anatomical neighbor point pair \(\{p,q\}\) with \(R(p,q)=1\), we set \(\varvec{L}[c,p]=1\) and \(\varvec{L}[c,1]=-1\).
Here we introduce a spatial-anatomical regularization instead of a spatial only regularization for two reasons: (1) voxel belong to different anatomical region may have different aging effect; (2) with SAR, we can divide all voxelwise features into several groups and learn the parameters for each group independently. This way can decrease the GPU memory cost and accelerate the computation by parallel computing.
2.3 Optimization and Implementation
With the new defined SAR penalty, the optimal voxelwise parameters are computed using:
where \(\lambda \) is a non-negative hyper-parameter to control to contribution of the voxelwise data fitting term and the SAR penalty. In our application, \(A(x,t_i)\) is equal to the input voxelwise feature map \(F_i(x)\). With the voxelwise parameter \(\varvec{\theta }(x)\), we can predict the voxelwise feature for a given age t, as \(Y(x,t)=f(t;\varvec{\theta }(x))\).
In this paper, the proposed method is implemented using Python and the GPU-based MxNet. In this implementation, we use the autograd functionality to compute the gradient with respect to the voxelwise parameters. It is easy to extend to other models for new applications. The cost function (Eq. 7) is optimized by ADAM optimizer [10], with 1000 iteration. The code will be public available for research usage.
3 Dataset
In this paper, we use the T1-weighted MR scans of the normal subjects from public available ADNI dataset (http://adni.loni.usc.edu/), up to Nov 27th, 2017. All selected normal subjects must have a CDR score equal or less than 0. The selected scans are divided into a training and a test group according to the ADNI study phase. The scans belong to ADNI 2 and ADNI 3 are used as training set, while the subjects in ADNI 1 and not in the first two year are treated as the test set. Overall, 609 scans were selected, including 430 scans for the training set and 179 scans for the test set. The demographic characteristics of the selected scans are summarized in Table 1.
All images are B1 corrected and preprocessed by a SPM-based Computational Anatomy Toolbox (CAT12 http://www.neuro.uni-jena.de/cat/) with the default parameter in the cross-sectional pipeline to generate gray matter and white matter density map. We use the neuromorphometrics atlas defined at the CAT12 template space as our segmentation \(\varvec{S}\), which contains 140 ROIs.
4 Experiment and Results
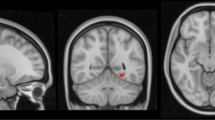
In the experiment, we first trained the voxelwise linear model with (\(\lambda >0\)) and without (\(\lambda =0\)) SAR penalty. For each method, the voxelwise parameters were learned using the training set on gray matter density feature map, and then the voxelwise gray matter map was predicted for the test set. For each ROI, the mean square error (MSE) between the observed voxelwise gray matter density feature F and the estimated one Y is computed. The difference of two methods is shown in Fig. 1. It is clear to see that, for each ROI on the test set, the linear model with SAR has smaller MSE than the linear model without SAR. The slope parameter (\(\varvec{\theta }_1\)) of the linear model learned with and without SAR overlaid on the T2 images, is shown in Fig. 2. As we can see, the slope parameter learned from linear model without SAR is very noisy and with no interpretability.
After verifying the effect of SAR penalty, the optimal parameter maps using general logistic model with SAR was learned on the gray density feature map. The barplot of the differences of MSE between two models in each ROI on the test set gray matter density feature is shown in Fig. 3. In the barplot, we found that there is only 1 ROI that linear model with SAR has smaller MSE compared to the general logistic model with SAR. Overall, the general logistic model with SAR beat the linear model on the rest 139 ROIs.
Beside the gray matter density map, we also learned the optimal parameter map using the general logistic with SAR on the white matter density map. When comparing the parameter maps from gray matter and white matter density, we found some interesting points. From Fig. 4, the parameter map (\(T_0(\varvec{\theta }_3)\) in the general logistic model), we found that the Caudate and Pallidum regions have smaller \(T_0\) in gray matter, which indicates the gray matter changes first occurred in these regions in normal aging process. The growth rate parameter \(\alpha (\varvec{\theta }_4)\) maps of gray matter and white matter were plotted in Fig. 5. In both maps, the positive value indicates the voxelwise feature increase while the negative value indicates the opposite effect. We can see that both gray matter and white matter features decreased with time in most brain regions. However, the voxelwise gray matter density tended to increase in regions around ventricle, which may be due to the expansion of ventricle. In both gray matter and white matter, large negative values were observed in the hippocampus region or around, which may indicate a fast degeneration of gray and white matter in these regions during aging process.
5 Conclusion
In this paper, we present a new method to model the aging related changes in voxelwise feature of the brain. It uses the voxelwise 4 parameters general logistic model and a spatial-anatomical regularization penalty term. The cost function is optimized using MxNet, which makes the method easy to extend to other applications. In the given voxelwise feature maps derived from ADNI, our method obtains a better voxelwise feature estimation. More important, our method offers a better interpretability of the learned parameter maps, which may help the clinician to learn the normal aging process from big data of normal population. Currently, only the voxelwise feature derived from T1 weighted MR image was used. It is of great interest to use our approach with multiple types of voxelwise features from different modalities and with starting points from different regions to indicate the aging or disease progression.
References
Sabuncu, M.R., Konukoglu, E.: Clinical prediction from structural brain MRI scans: a large-scale empirical study. Neuroinformatics 13(1), 31–46 (2015)
Stonnington, C.M., Chu, C., Klppel, S., Jack, C.R., Ashburner, J., Frackowiak, R.S.: Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. NeuroImage 51(4), 1405–1413 (2010)
Frisoni, G.B., Fox, N.C., et al.: The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 6(2), 67 (2010)
Rao, A., Monteiro, J.M., Mourao-Miranda, J.: Predictive modelling using neuroimaging data in the presence of confounds. NeuroImage 150, 23–49 (2017)
Gould, I.C., Shepherd, A.M., et al.: Multivariate neuroanatomical classification of cognitive subtypes in schizophrenia: a support vector machine learning approach. NeuroImage Clin. 6, 229–236 (2014)
Rao, A., Monteiro, J.M., et al.: A comparison of strategies for incorporating nuisance variables into predictive neuroimaging models. In: 2015 International Workshop on Pattern Recognition in NeuroImaging (PRNI), pp. 61–64. IEEE (2015)
Dukart, J., Schroeter, M.L., Mueller, K., et al.: Age correction in dementia-matching to a healthy brain. PLoS One 6(7), e22193 (2011)
Tao, Q., Lamb, H.J., Zeppenfeld, K., van der Geest, R.J.: Myocardial scar identification based on analysis of look-locker and 3D late gadolinium enhanced MRI. Int. J. Cardiovasc. Imaging 30(5), 925–934 (2014)
Cuingnet, R., Glaunès, J.A., Chupin, M., Benali, H., Colliot, O.: Spatial and anatomical regularization of SVM: a general framework for neuroimaging data. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 682–696 (2013)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Sun, Z., Xu, W., Wang, S., Xu, J., Qiao, Y. (2018). Modeling Longitudinal Voxelwise Feature Change in Normal Aging with Spatial-Anatomical Regularization. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11072. Springer, Cham. https://doi.org/10.1007/978-3-030-00931-1_46
Download citation
DOI: https://doi.org/10.1007/978-3-030-00931-1_46
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00930-4
Online ISBN: 978-3-030-00931-1
eBook Packages: Computer ScienceComputer Science (R0)