Abstract
The local spatial arrangement of nuclei in histopathology image has been shown to have prognostic value in the context of different cancers. In order to capture the nuclear architectural information locally, local cell cluster graph based measurements have been proposed. However, conventional ways of cell graph construction only utilize nuclear spatial proximity, and do not differentiate different cell types while constructing a cell graph. In this paper, we present feature driven local cell graph (FeDeG), a new approach to constructing local cell graphs by simultaneously considering spatial proximity and attributes of the individual nuclei (e.g. shape, size, texture). In addition, we designed a new set of quantitative graph derived metrics to be extracted from FeDeGs, in turn capturing the interplay between different local cell clusters. We evaluated the efficacy of FeDeG features in a digitized H&E stained tissue micro-array (TMA) images cohort consists of 434 early stage non-small cell lung cancer for predicting short-term (<5 years) vs long-term (>5 years) survival. Across a 100 runs of 10-fold cross-validation, a linear discriminant classifier in conjunction with the 15 most predictive FeDeG features identified via the Wilcoxon Rank Sum Test (WRST) yielded an average of AUC = 0.68. By comparison, four state-of-the-art pathomic and a deep learning based classifier had a corresponding AUC of 0.56, 0.54, 0.61, 0.62, and 0.55 respectively.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
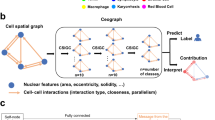
1 Introduction
Changes in distribution, appearance, size, morphology, and arrangement of histologic primitives, e.g., nuclei and glands, can predict tumor aggressiveness. For instance, in the context of lung cancer, it is known that more and less aggressive diseases are characterized by differences in nuclear shape, morphology and arrangement. For a number of different cancers, the hallmark of presence of disease is the disruption in the cohesion of architecture between nuclei and other primitives belonging to the same family, e.g. nuclei or lymphocytes. Conversely, aggressive tumor tends to exhibit relatively lower degrees of structure and organization between the same class of primitives compared to less aggressive cancers.
There has been recent interest in developing computational graph-based approaches to characterize spatial arrangement of nuclei in histopathology images to be able to predict patient outcome [1,2,3]. Many of these approaches are based on global graphs such as Voronoi and Delaunay triangulation strategies to connect individual nuclei (representing graph vertices or nodes) and then computing and associating statistics relating to edge length and node density to disease outcome. Lewis et al. proposed cell cluster graphs (CCG) in which the nodes are defined on groups/clusters of nuclei rather than on individual nucleus [3], since there is a growing recognition that tumor aggressiveness might be driven more by the spatial inter-actions of proximally situated nuclei, compared to global interactions of distally located nuclei. While these approaches showed that attributes relating to spatial arrangement of proximal nuclei were prognostic [1,2,3], the graph connections did not discriminate between different cell populations, e.g. whether the proximal cells were all cancer cells or belonged to other families such as lymphocytes.
In this work we seek to go beyond the traditional way of constructing cell graphs, which focus solely on cell proximity. Instead we seek to incorporate the intrinsic nuclear morphologic features coupled with spatial distance to construct locally and morphologically distinct cell clusters. We introduce a new way of constructing a local cell graph called the Feature Driven Local Cell Graph (FeDeG), along with a corresponding new set of quantitative histomorphometric features. In the context of early stage lung cancer, we demonstrate that FeDeG features extracted from nuclear graphs from digitized pathology images are predictive of overall survival.
2 Brief Overview and Novel Contributions
The novel contributions of this work include: (1) FeDeG is a new way to construct local cell graphs based on the nuclear features. This results in locally packed cell graphs comprising nuclei with similar phenotype. Figure 1 illustrates the global cell graph [1], CCG [3] and FeDeG graph in the same local region contains lymphocytes and cancer cells. One may see that the global graph connects all the nuclei in the image may not allow for capturing of local tumor morphology efficiently. Similarly, the CCG only considers nuclear locations which results in connecting lymphocytes and cancer nuclei into a graph, important information involving local spatial interaction between different cellular clusters may be left unexploited.
The FeDeG incorporates nuclear morphologic feature (nuclear mean intensity in this case) into the graph constructing process, which enable us to interrogate the interaction between different graphs and to reveal more sub-visual information from the underlying tissue image.
(2) In addition to construct local cell graphs using FeDeG, we designed a new set of quantitative histomorphometrics based on the constructed FeDeGs. These features are: Intersection between different FeDeGs, size of FeDeGs, Disorder of nuclear morphology within FeDeG and the spatial arrangement of FeDeG. There features are different compared to standard features extracted from CCG and global graph methods, which only quantify the density of local/global graph, or the local/global distances between cells. The FeDeG features attempt to capture the interactions between and within local cell clusters with similar morphological properties.
(3) We employ the FeDeG and associated quantitative features in conjunction with a linear machine learning classifier to predict risk of recurrence in early stage non-small cell lung cancer (NSCLC). Similar work has been reported by Yu et al. [4] and Wang et al. [5] for predicting recurrence in early stage NSCLC patients, in which the global architecture and shape of nuclei features were found to be predictive. However, the interactions between different local cell clusters have not been explored. In the experiment, we compared the FeDeG with these nuclear features. Figure 2 shows the flowchart of FeDeG construction and associated feature computation, which include nuclei segmentation, FeDeG construction, and FeDeG feature extraction modules.
Flowchart of FeDeG construction and associated feature computation. (a) Green contours indicate the nuclear boundaries (b) FeDeG construction based on the nuclear features (used mean intensity of nuclei in this case). Nuclei that belong to the same FeDeG have connecting edges with the same color. (c) FeDeG feature computation. (Color figure online)
3 Feature Driven Cell Graph
3.1 Nuclei Segmentation and Morphologic Feature Extraction
In order to efficiently segment nuclei a multiple-pass adaptive voting method was employed to detect the cells [6] followed by a local optimal thresholding method that segments nuclei based on analyzing the shape of these cells as well as the area occupied by them. A set of 6 nuclear morphologic features that described the nuclear shape, size and texture were then computed based on these pre-segmented nuclei.
3.2 FeDeG Construction in Nuclear Morphologic Feature Space
In this step, spatial and morphological features of nuclei were used for feature space analysis to construct FeDeG. The mean-shift clustering [7] was applied to perform the feature space analysis for sub-graph construction. It accomplishes this by first estimating the modes (i.e., stationary points of the density of nuclear morphologic feature) of the underlying density function of the nuclear morphologic feature. It then groups nuclei into different sub-graphs based on the corresponding modes.
We denote as N the total number of nuclei in the image, and each nucleus has a corresponding feature vector in d-dimensional Euclidean space \( \varvec{R}^{d} \), so that we have a set of nuclear feature vectors \( \varvec{X = x}_{1} ,\;\varvec{x}_{1} ,\; \cdots ,\;\varvec{x}_{N} ,\;\text{where}\;\varvec{x}_{n} \in \varvec{R}^{d} \). For each feature vector \( \varvec{x}_{n} \in \varvec{X} \), there is a corresponding mode \( \varvec{y}_{i} \). In the beginning, the mode \( \varvec{y}_{i} \) is initialized with the original feature vector \( \varvec{x}_{n} \) i.e., \( \varvec{y}_{i}^{o} = \varvec{x}_{n} \). The \( \varvec{y}_{i}^{u} \) is then recursively updated, based on the neighborhood nuclear characteristics, using the following equation:
where \( \varvec{y}_{i}^{u + 1} \) is the updated version of \( \varvec{y}_{i}^{u} \). The vector \( \varvec{m}_{G} (\varvec{y}_{i}^{u} ) \) is called the mean-shift vector and calculates the difference between the weighted mean and the center of the kernel. It has been previously shown that the mean-shift vector always points toward the direction of maximum increase in the underlying density function [Comaniciu2002]. At the final step, each nuclear feature vector \( \varvec{x}_{n} \) finds a corresponding mode \( \varvec{y}_{i} \) which will be used for constructing the FeDeG.
In this work, we explored a Q-dimensional feature space which includes 2-D spatial coordinates (i.e., centroid location) of nuclei in the image and Q-2 of the nuclear morphologic features. These features are chosen based on the observation that the same types of nuclei are usually located closely together and have a similar phenotype. The corresponding multivariate kernel is defined as the product of two radially symmetric kernels as follows:
where k(·) is the profile of the kernel, xs is the spatial component, xm is the nuclear morphologic component, C is the normalization constant, and hs and hm are the kernel bandwidths controlling the size of the kernels. The higher value of the kernel bandwidths hs and hm correspond to more neighboring data points that are used to estimate the density in the Q-D feature space. This can be seen in Fig. 2(d), in which the FeDeGs were constructed in a 3-D feature space (2D coordination + nuclear intensity).
3.3 FeDeG Features Computation
Based on the FeDeG created, four groups of features were derived as show in Table 1. These quantitative features were aiming to measure the interaction between FeDeGs, intrinsic nuclear variation within each FeDeG, and spatial arrangement of FeDeGs.
4 Experimental Design
4.1 Dataset Description
The early stage NSCLC cohort comprises a total of 434 patients in the form of digitized TMA image (scanned at 20X magnification digitally). Long term clinical outcome was available for all patients in this cohort (collected between 2004 and 2014), which ends up with 280 short-term survival patients (<5 years after surgery) and 154 long-term survival patients (>5 years after surgery).
4.2 Comparative Methods
4.2.1 Graph Based and Other Pathomic Strategies
The goal of the experiment is to be able to predict short vs. long term survival in NSCLC patients. By doing that, we built separated predictive models based on pathomic features extracted from the histologic TMA spots. We compared the efficacy of FeDeG features with four states-of-the-art histomorphometric based approaches involving description of cell morphology and architecture which has been reported to have prognostic values [3,4,5, 8]. For all the feature sets, the nuclear segmentation from Sect. 3.1 was used to calculate the nuclear boundaries and centroids. In total, we investigated the performance of 5 feature sets: (1) 100 features describing nuclear shape [4, 5], (2) 51 features describing global cell architectures [5], (3) 24 features describing cell orientation entropy by COrE [8], (4) 35 CCG features describing local cell cluster arrangement [3], and (5) 176 FeDeG features describing the interaction between local cell clusters comprising nuclei with similar properties. A linear discriminant analysis classier (LDA) was implemented and trained based on the patient labels for samples, under 10-fold cross-validation (CV) with 100 runs. Within each fold, top 10 predictive features for each of the 5 feature groups were selected by using Wilcoxon rank sum test (WRST). The mean area under the receiver operating characteristic curve (AUC), was used to evaluate and compare the different classifiers.
4.2.2 Deep Learning
We also compared the FeDeG features with a deep learning method (DLM). The DLM was implemented using the Alexnet style Convolutional Neural Network (ConvNet). Specifically, a 10-layer ConvNet architecture comprising 1 input layer, 5 convolution layers, 3 fully connected layers and 1 output layer was constructed. The input layer accepts an image patch of 256 × 256 pixels, and the output layer is a soft-max function which outputs the class probability of being positive or negative. In the DLM, we split each TMA spot image into smaller patches of 200 × 200 pixels, the class labels for these image patches being assigned the same class label as that of the corresponding TMA spot image it was derived from. The average image size of the TMA spot was 3000 × 3000 pixels at 20× magnification, which in turn resulted in a total number of about 68,000 patches after filtering out unusable patches. We performed the training and testing using a 10-fold cross-validation approach across each fold, all training and testing being done at the patient and not at the individual image-level. Once each of the individual image patches corresponding to a single patient have been assigned a class label, majority voting was employed to aggregate all the individual predictions to generate a patient-level prediction.
5 Results and Discussion
5.1 Discrimination of Different Graph and Deep Learning Representations
Figure 3(b) show the classification performance comparison based on different types of feature sets. The FeDeG based classifier achieved the highest AUC of 0.68 ± 0.02, outperforming the other feature sets. The Global graph, shape, COrE, CCG, and DL feature classifiers respectively yielded AUCs of 0.56 ± 0.02, 0.54 ± 0.03, 0.61 ± 0.02, 0.62 ± 0.03, and 0.55 ± 0.04, respectively. The Receiver operating characteristic (ROC) curves are shown in Fig. 3. The classification results suggested that the locally extracted nuclear features provided better prognostic value than those associated with global architecture. Comparing the performance of CCG and FeDeG based classifiers suggests that the organization of local cell clusters, where cluster membership was defined not solely based off spatial proximity but also on morphologic similarity resulted in more prognostic signatures. Figure 4 shows two representative H&E stained TMA spot images for long-term and short-term survival NSCLC patients along with the corresponding CCG and FeDeG feature representations. The panel inset for FeDeG reveals the grouping of the TIL and cancer nuclei as distinct clusters with associated spatial interaction between these two cell families, unlike the CCG representation which does not distinguish between the nuclei and TILs.
(a) The box-plot of top-2 FeDeG features in distinguishing short vs. long-term survival. (b) The ROC curves for all comparative strategies. (c) The KM curves for the FeDeG classifier associated with short and long-term survival under a leave-one-out framework (p = 0.00772, HR (95% CI) = 1.59(1.15–2.21)).
Representative H&E stained TMA spot images for (a) long-term survival and (d) short-term survival NSCLCs with CCG and FeDeG feature representations. The second column shows the CCG configuration. The third column shows the FeDeG configuration, in which FeDeG graphs were colored. The panel inset for FeDeG reveals the grouping of the TIL and cancer nuclei as distinct clusters with a spatial interaction between these two cell families, unlike the CCG representation which does not distinguish between them.
Building a good deep learning model normally requires a large amount of well-annotated training cases. The deep learning approach we employed was constrained by the fact that we had an unbalanced dataset, (280 short-term vs. 154 long-term survivals). It is likely that the relatively few long-term survival patients, coupled with the class imbalance resulted in a sub-optimally trained deep learning network. Also, while DLMs have been reported good at low-level visual object detection and segmentation tasks, it is still unclear now how to use DLMs to summarize the sub-visual information extract from image patches in order to make prognostic predictions.
During feature discovery, we found that measures of the degree of FeDeGs intersection, and the variance of FeDeG graph sizes were the most two frequently selected features by WRST across 100 runs of 10-fold cross-validation (the boxplot of these two FeDeG features are shown in Fig. 3(a)). The top 1 feature reflects the degree of interactions between different local cell families. The boxplot in Fig. 3(a) appears to suggest that tumor outcome is improved with an increase in the total number of local cell cluster interactions. This may in turn reflect increase spatial interplay between tumor infiltrating lymphocytes (TIL) and cancer nuclei clusters. This is also reflected in the FeDeG maps shown in Fig. 4(c) and (f), in which we observe a higher number of intersections between nearby FeDeG graphs in the case of patient with long-term survival (Fig. 4(c)), compared to a short-term survivor (Fig. 4(f)).
5.2 Survival Analysis
In univariate survival analysis, the Log-rank test was performed based on the predicted labels generated by FeDeG classifier. The patients identified as high risk had significantly poorer overall survival, with Hazard Ratio (95% Confident Interval) = 1.59 (1.15–2.21), p = 0.00672. We set the threshold for statistical significance at 0.05, and none of the other comparative were found to be statistically significantly prognostic of survival in NSCLC.
6 Concluding Remarks
We presented a new approach called feature driven local cell graph (FeDeG), which provide a new way to construct local cell graph. A new set of histomorphometric features also derived based on the constructed FeDeGs, the aim of which was to quantify the interaction and arrangement of local cell cluster comprising of nuclei with similar properties. The FeDeG feature based classifier showed a strong correlation with overall survival in non-small cell lung cancer patients and yield superior classification performance compared to the other pathomics. Going forward, we will attempt to validate FeDeG in larger cohorts and other tissue types, such as breast cancer.
References
Bilgin, C., et al.: Cell-graph mining for breast tissue modeling and classification. In: International Conference on IEEE Engineering in Medicine and Biology Society, pp. 5311–5314. IEEE (2007)
Shin, D., et al.: Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin. Gastroenterol. Hepatol. 13, 272–279.e2 (2015)
Lewis, J.S., et al.: A. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am. J. Surg. Pathol. 38, 128–137 (2014)
Yu, K.-H., et al.: Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 7(12474), 1–10 (2016)
Wang, X., et al.: Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci. Rep. 7(1), 13543 (2017)
Lu, C., et al.: Multi-pass adaptive voting for nuclei detection. Sci. Rep. 6(1), 33985 (2016)
Comaniciu, D., Meer, P.: Mean shift: a robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 24, 603–619 (2002)
Lee, G., et al.: Cell orientation entropy (COrE): predicting biochemical recurrence from prostate cancer tissue microarrays. In: MICCAI, pp. 396–403 (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Lu, C. et al. (2018). Feature Driven Local Cell Graph (FeDeG): Predicting Overall Survival in Early Stage Lung Cancer. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_46
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_46
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)







