Abstract
Breast cancer is the most common cancer in women, and ultrasound imaging is one of the most widely used approach for diagnosis. In this paper, we proposed to adopt Convolutional Neural Network (CNN) to classify ultrasound images and predict tumor malignancy. CNN is a successful algorithm for image recognition tasks and has achieved human-level performance in real applications. To improve the performance of CNN in breast cancer diagnosis, we integrated domain knowledge and conducted multi-task learning in the training process. After training, a radiologist visually inspected the class activation map of the last convolutional layer of trained network to evaluate the result. Our result showed that CNN classifier can not only give reasonable performance in predicting breast cancer, but also propose potential lesion regions which can be integrated into the breast ultrasound system in the future.
J. Liu and W. Li — Have equal contribution.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Breast cancer is the most common cancer in women. According to statistics in 2013, breast cancer caused approximately 8.2–14.94 million death worldwide [2]. The corresponding morbidity rate is 15.1%, while the death rate is about 6.9%. Detection and diagnosis in the early stage are essential for its treatment, which could improve survival rate. One of the most efficient diagnostic methods is mammographic screening. However, mammographic sensitivity can be relatively low in dense breasts (less than 50%) [2], which may lead to unnecessary breast biopsies (65–85%) [3]. Whereas, with the interpretation of skilled radiologists, ultrasonography presents a higher accuracy in distinguishing benign breast lumps from malignant tumors. A US population study has demonstrated that by using brightness mode ultrasound, the overall sensitivity could reach 97.2% (281 of 289). The specificity can achieve 61.1% (397 of 650) and the accuracy could be 72.2% (678 of 939) [4]. In addition, ultrasound is radiation free, easily accessible, economical and convenient in practice. Therefore, ultrasonography has gradually become an alternative to mammography in clinical diagnosis of breast cancer.
The Breast Imaging Reporting and Data System (BI-RADS) [5] offered standardized terminology to depict features, and to provide assessments as well as recommendations. Features including shape, orientation, margin, echo pattern, and posterior features of masses are compiled in the BI-RADS lexicon for ultrasound. Based on the particular features of the lesion, radiologists would recommend one BI-RADS category (Table 1). Radiologists would finally issue a clinical recommendation according to these categories, which suggested an annual examination for categories 1 and 2, an extra test six months later for category 3, and a biopsy for categories 4, 5 and 6 [6]. But the image-based diagnosis was dependent on practitioners’ experience and thus relatively subjective. A computer aided diagnosis system is of great demands to resolve this issue.
Thanks to the emerging deep learning technique such as convolutional neural network (CNN) [7], computer aided automatic detection system can now achieve comparable or even better performance than radiologists in detecting lesion or diagnosing medical conditions from image data. For instance, with CNN, computers achieved 90% sensitivity and 85% specificity in predicting brain hemorrhage, mass effect, or hydrocephalus from CT images [8], successfully predicted diabetic retinopathy patients among 11711 retinal fundus photographs with 96% sensitivity and 93% specificity [9], or even can diagnose skin cancer with only photos taken by smartphone [10]. In previous works, based on CNN, breast cancer diagnosis systems have also been proposed to classify breast cancer in mammogram images [11] or segment breast tumor regions in histopathological images [12]. Thus, this paper seeks feasibility of utilizing CNN to predict breast cancer in ultrasound images into one of the three malignancy categories: malignant tumor, benign lump, or normal tissue. We adopted BIRADS categories as a “domain knowledge” to improve the classification of ultrasound images. Specifically, two different classifiers have been proposed. One directly classifies the malignancy and the other one simultaneously predicts BI-RADS category. By this means, BIRADS categories, which can be interpreted as doctors’ visual interpretation of image features, can guide the training of image features obtained by CNN to improve the performance of malignancy claasification. The performance of each classifier and the cause of failures were then examined in details. We will show that CNN classifier can not only give reasonable performance in predicting breast cancer but also propose potential lesion regions.
2 Method
2.1 Data Acquisition
We retrospectively collected 1933 breast ultrasound images from 608 patients. Eligibility criteria excluded patients who received breast implants or surgeries on the ipsilateral breast, and those who were pregnant or breastfeeding. All images were reviewed by experienced radiologists and BI-RADS assessments [5] were recorded (Table 1). Based on BI-RADS categories, images were then classified as malignant tumor (BI-RADS categories 5, 6, and part of 4), benign lump (BI-RADS categories 2, 3 and part of 4), and normal tissue (BI-RADS category 1). Since radiologists cannot directly verify the malignancy of lumps classified as BI-RADS category 4 with ultrasound images, a pathological test was conducted by ultrasound-guided core needle biopsy (CNB) or a surgery following the clinical procedures to confirm the malignancy. In total, 96 BI-RADS category 4 images were classified as malignant. To summary, among the images acquired, 491 show malignant tumor, 426 show benign lump, and 1016 are normal tissues.
2.2 Image Preprocessing
After collecting data, we pre-processed the data following the steps below:
Cropping Images.
The ultrasound images collected had uninformative parts, such as screen background, acquisition parameters and hospital name. Since these are not helpful and may even introduce bias, we manually cropped all the images to remove them. In addition, since the shape of ultrasound images may vary across devices and imaging settings – some are in horizontal rectangle while others are in vertical rectangle shape, we cropped the images into square shape for the training convenience. This may remove useful information in the image. To avoid that, we ensured to keep the complete lump which is critical for the prediction, and the skin tissues whose contrast is more accurate than deep tissues and more uniform across devices.
Removing Makers:
In some ultrasound images radiologists left markers to indicate or quantify tumor position (e.g. cross symbol, rectangle box, dash line). These markers, colorized or black-and-white, may introduce bias to the training of classifier. For instance, classifier may likely take cross symbols as a tumor indicator since no such symbols appear on normal tissues. Hence, we adopted a semi-automatic approach to clean those symbols. Considering that the ultrasound images we collected are in gray color, we first detected all the colored or pure black-or-white pixels in the image as a mask of potential markers. The mask was reviewed and cleaned manually with in-house image annotation tools. Then, the masked pixels were restored by linear interpolation with the surrounding pixels. All images were reviewed after processing to ensure that no obvious artifacts were left and the failed ones were excluded.
2.3 Training Multi-task CNN
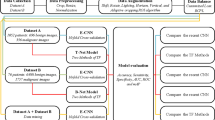
We adopted the convolutional layers in VGG16 [7] to extract image features for the classifier. VGG16 contains 13 convolutional layers and 5 max pooling layers. It is a well-established CNN classifier for image recognition tasks and has been utilized in many applications. Though there are other deeper and more advanced CNN classifiers, we decided to choose VGG16 to avoid potential overfitting issues considering the limited training data in our experiments. Following this philosophy, a dense layer that is small than the original VGG16 network was cascaded after flattened convolutional layers (Fig. 1).
When training the classifier, we applied the weights pre-trained based on ImageNet database as the initial weights. Notably, since our ultrasound images are monochrome but pre-trained VGG16 network takes colorful images with 3 channels as input, we need to first convert the monochrome image to colorful images. One typical approach that is widely used by others is to use the same image in all 3 channels. However, in our view, the redundant information introduced in those approaches does not fully embrace the power of CNN. Instead, we inserted a convolutional layer with three 3 × 3 kernels between the input layer and VGG16 convolution layers. This layer will be trained to convert input monochrome images into the three channel images that best fits the pre-trained VGG16 network.
In our baseline method, the network proposed above only classified the image into 3 classes: malignant tumor, benign lump, and normal tissue. We wondered if the performance of this task can be further improved by introducing clinical domain knowledge into the training process. Thus, when training the network, another logistic regression classifier was appended after the convolution layers to classify the image into the BI-RADS categories (Fig. 1). Though this might add an extra burden to the network and BI-RADS categories are highly coupled with tumor prediction, our rationale of this design is as follows: (1) BI-RADS assessment is based on professionals’ visual inspection of the image features. It might help guide the training of the tumor related features. (2) BI-RADS categories are finer than the 3 maliginacy classes we want to predict and thus may offer better guidance. (3) Additional burden in the training process may help to reduce the chance of overfitting.
3 Results
In our experiments, data was evenly split into five folds. Since multiple ultrasound images may be acquired for each patient, we ensured that the images from the same patient will be assigned to the same fold. Also, we tried to keep the distribution of each class as the same as possible in each fold. To fully utilized the data to examine our proposed method, the training process follows cross-validation training scheme – each time take four folds as training set and test on the rest fold. Five independent trainings and testings were conducted on both the baseline network and the multi-task network.
3.1 Classification Result
We combined the testing results of five independent experiments and conducted quantitative analyses accordingly. Table 2 shows the number of images in each category. Though the result is not perfect, both classifiers worked reasonably well in classifying images. With the baseline method, the prediction accuracy is 82.9%. By using the multi-task network, the prediction accuracy slightly increased to 83.3%. Notably, both methods have high sensitivity in differentiating abnormal cases from normal ones (baseline: 95%, multi-task: 96.7%). The major error comes from separating malignant tumors and benign lumps. For the proposed multi-task approach, only 74.3% malignant (baseline: 71.9%) tumors were correctly classified. This is reasonable since the malignancy level of BI-RADS category 4 tumor is also difficult for experts to tell based on an ultrasound image only. About half of the errors between malignant tumor and benign lump prediction happened in BI-RADS category 4 images (baseline: 57.5%, multi-task: 54.9%). Overall, despite the reduced sensitivity in predicting normal tissues (baseline: 93.7%, multi-task: 91.2%), better performance has been achieved by the proposed multi-task approach.
3.2 Examples of Correct and Wrong Predictions
To further examine the performance of each classifier and understand why this classification task is challenging, we selected some example images. In order to understand what happened inside the CNN classifier, class activation map (CAM) of the last convolutional layer was visualized [13]. CAM is a heat map highlights the attention of a classifier when making the decision and thus can reveal the regions associated with the prediction. Specifically, two groups of examples were selected and shown. (1) The tumors/lumps correctly predicted by the multi-task method only and the normal tissues correctly predicted by the baseline method only were shown (Fig. 2). (2) A considerable number of images were wrongly classified by both methods, examples of those images were shown (Fig. 3). The images and the corresponding CAM were examined by experienced radiologists.
As shown in Fig. 2a–d, when the classification is correct, CAM accurately highlighted the malignant masses (Fig. 2a–b) and benign lumps (Fig. 2c–d). When the baseline method wrongly classified malignant tumors as benign, the network pays attention to both CAM area as well as the adjacent normal tissues. As for the normal cases which were correctly predicted by the baseline only, the attention is on the normal shallow skin tissues while the multi-task method wrongly regarded the decay resulted from deep location or dense superficial tissues and some cellulite that extended into the glandular layer as masses (Fig. 2e–f).
As for the cases failed by both methods, some of them belongs to BI-RADS category 4, which is also difficult for radiologists to decide and requires pathological verifications (Fig. 3b–c). Nevertheless, for these cases, CAM still accurately highlighted lumps in the image. Figure 3d was predicted as normal tissues due to the tiny volume of the mass. Some radiologists may consider it as normal ducts while others may think of cysts. The interpretation on this kind of images is relatively subjective, and the lesion has little impact on patients. In Fig. 3e, a malignant label was given to normal tissues, as the area was a centralized point of mammary ducts and thus was difficult to be distinguished from lesions even by senior doctors, if the location is unclear. Another misdiagnosis example was Fig. 3f, in which normal tissues were deemed as benign lumps. The interpretation identified the cellulite as a hypoechoic mass. It was difficult to tell the exact nature of this mass, since ultrasound radiologists also depended on whether the mass continued with normal tissues to determine its character. Due to the complex structures of breasts, such as the cellulite penetrating into the glands, the collection of vasa efferentia under the nipples, the common features of benign and malignant lumps, together with the different features of each section, it is difficult to determine the relationship between lumps and its surrounding tissues, as well as its overall situation. More complete patient-based videos might be required to obtain better results.
Notably, in our preliminary results, we found some intriguing malignant cases that were classified as normal (e.g. Fig. 3a). After reviewing the cases, we found that some of them came from the patients which were diagnosed as cancer. But the tumor was not captured in the image and the tissues shown in the image are normal findings. Those cases were eliminated with a second review. This also suggests that CNN is a powerful tool to learn the generalized pattern even when there are noises in the training data.
4 Conclusion
In this paper, we adopted CNN to predict breast tumors in ultrasound images. Domain knowledge was integrated into the training process. Promising results were obtained in separating images with lump and normal findings. Moreover, though this is not a segmentation task, the activation map of the trained classifier can still correctly highlight the mass regions in images. In addition, reasonable results were also obtained in differentiating malignant tumors and benign lumps. In the future, more data will be collected to fine-tune the network. And the system will be extended to process video data for better classification and prediction of the tumor malignancy. The correlation between BI-RADS categories and the classification results will be then investigated such that the whole system can be integrated into current breast cancer diagnosis procedure.
References
Fitzmaurice, C., et al.: The global burden of cancer 2013. JAMA Oncol. 1(4), 505 (2015)
Kolb, T.M., Lichy, J., Newhouse, J.H.: Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology 225(1), 165 (2002)
Jesneck, J.L., Lo, J.Y., Baker, J.A.: Breast mass lesions: computer-aided diagnosis models with mammographic and sonographic descriptors. Radiology 244(2), 390 (2007)
Berg, W.A., et al.: Shear-wave elastography improves the specificity of breast US: the BE1 multinational study of 939 masses. Int. J. Med. Radiol. 262(2), 435 (2012)
Mendelson, E., et al.: Breast imaging reporting and data system, BI-RADS: ultrasound (2003)
D’Orsi, C., et al.: ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System (2013)
Simonyan, K., Zisserman, A.: Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv:1409.1556 (2014)
Prevedello, L.M., et al.: Automated critical test findings identification and online notification system using artificial intelligence in imaging. Radiology 285(3), 162664 (2017)
Gulshan, V., et al.: Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316(22), 2402 (2016)
Esteva, A., et al.: Corrigendum: dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115 (2017)
Sun, W., et al.: Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput. Med. Imaging Graph. 57, 4–9 (2017)
Su, H., et al.: Region segmentation in histopathological breast cancer images using deep convolutional neural network. In: IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 55–58 (2015)
Kotikalapudi, Raghavendra and contributors: https://github.com/raghakot/keras-vis (2017)
Acknowledgement
The work received supports from Shenzhen Municipal Government under the grants JCYJ20170413161913429 and KQTD2016112809330877.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, J. et al. (2018). Integrate Domain Knowledge in Training CNN for Ultrasonography Breast Cancer Diagnosis. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_96
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_96
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)