Abstract
Accurate gross tumor volume (GTV) segmentation in esophagus CT images is a critical task in computer aided diagnosis (CAD) systems. However, because of the difficulties raised by the contrast similarity between esophageal GTV and its neighboring tissues in CT scans, this problem has been addressed weakly. In this paper, we present a 3D end-to-end method based on a convolutional neural network (CNN) for this purpose. We leverage design elements from DenseNet in a typical U-shape. The proposed architecture consists of a contractile path and an extending path that includes dense blocks for extracting contextual features and retrieves the lost resolution respectively. Using dense blocks leads to deep supervision, feature re-usability, and parameter reduction while aiding the network to be more accurate. The proposed architecture was trained and tested on a dataset containing 553 scans from 49 distinct patients. The proposed network achieved a Dice value of \(0.73\pm 0.20\), and a 95\(\%\) mean surface distance of \(3.07 \pm 1.86\) mm for 85 test scans. The experimental results indicate the effectiveness of the proposed method for clinical diagnosis and treatment systems.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
One of the most critical challenges in radiotherapy (RT) treatment planning is a robust strategy for delineation of the gross tumor volume (GTV). Manual segmentation of the GTV is time consuming, subject to error, and involves valuable human resources. Hence, a great deal of effort has been devoted to automating the process for different organs in CT images. Esophageal cancer is the eighth common form of cancer worldwide with 456,000 new cases yearly, and the sixth most fatal form of cancer [1]. RT is one of the treatment options, both in palliative and curative settings. Delineation of the GTV is not trivial due to short and long term shape changes, and sometimes poor visibility on CT scans used for RT treatment planning. Therefore, physicians use a combination of clinical history, endoscopic findings, and other imaging modalities in conjunction with CT imaging for manual delineation of the esophageal GTV. Obtaining this data is hard, time consuming and expensive. Thus, developing an automatic and reliable esophageal GTV segmentation approach is desirable. However, automatic esophageal GTV segmentation in CT scans has been addressed rarely, and is much harder than segmenting the esophagus due to the difficulties raised by the versatile shape, the poor contrast of the tumor with respect to adjacent tissues, and the existence of foreign bodies in the esophageal lumen.
Lately, convolutional neural networks (CNNs) have attracted a great deal of attention for medical image analysis [2]. However, very few CNN segmentation techniques have been proposed in the context of esophagus segmentation and most of them are highly user interactive. Fechter et al. [3] proposed a fully CNN (FCNN) for segmenting the esophagus in 3D CT images. Because of poor visibility of the transition from the esophagus to the stomach, only the region between the lower tip of the heart and the upper side of the stomach was considered. An active contour model and a random walker were used as post-processing steps. The network achieved an average Dice value of 0.76 ± 0.11 for 20 test scans. A semi-automatic two-stage FCNN for 2D esophagus segmentation was proposed in [4], extracting an ROI in the first stage, and performing the segmentation in the second stage. A Dice value of 0.72 ± 0.07 was reported for 30 test scans. Hao et al. [5] used an FCNN as a pre-processing step for extracting a ROI in 2D CT scans. Then a graph cut method for segmenting the tumor was applied. An average Dice value of 0.75 ± 0.04 was reported for 4 test scans.
In this paper, we propose a 3D end-to-end CNN for esophageal tumor segmentation. The proposed architecture, called 3D-DenseUnet, is related to fully convolutional DenseNet (FC-DenseNet) [6, 7], but uses 3D convolutions rather than 2D. In this paper we leverage the idea of dense blocks, arranging them in a typical U-shape. This improves the flow of information and gradients throughout the network and strengthens feature propagation and feature re-usability. Different from [7], two techniques of bottleneck layers and feature compression are used in order to increase the feature maps in a tractable fashion. Also, we adapt the loss function particularly for our dataset. To the best of our knowledge this is the first end-to-end method that addresses automatic 3D tumor segmentation in esophageal CT scans of the whole chest region.
2 Proposed Network Architecture
The proposed DenseUnet, see Fig. 1, is a 3D network composed of a contractile path to extract contextual features and an expanding path to recover the input patch resolution. Each path consists of three main components: dense blocks, down-sampling units, and up-sampling units. Since memory usage in 3D CNNs is a challenging issue, training is performed using 3D patches rather than complete scans. The input patch size is \(47\times 47\times 47\) voxels, i.e. encompassing the GTV, while the output is probabilistic with size \(33\times 33\times 33\) voxels, concentric with the input. In the contractile path at each level, there is a dense block. The two first contractile levels are followed by down-sampling units which reduce the number of parameters and make the network capture more contextual information. At the third level of the network, a dense block and an up-sampling unit are stacked. Up-sampling layers and skip connections assist the network to retrieve the lost resolution after the down-sampling units. At the final step, the network is followed by one conv\((3\times 3\times 3)\)-BN-ReLU (where BN is batch normalization and ReLU a rectified linear unit), another convolutional layer with linear activation, and a soft-max layer in order to compute a probabilistic output which can be classified as GTV and background.
Figure 2 illustrates the structure of the main components of the proposed network. In each dense block, one conv\((1\times 1\times 1)\)-BN-ReLU and one conv\((3\times 3\times 3)\)-BN-ReLU are stacked. Dense blocks with direct connectivity between all the subsequent layers, improve the information flow between the layers and make the network more accurate [6]. Also, due to its feature reusing capability, dense blocks can perform deep supervision [7]. Unlike FC-Densenet, we employ a feature reduction technique to avoid feature explosion. In each dense block conv\((1\times 1\times 1)\) layers are used as bottleneck layers, which reduce the number of input feature maps and thus improve computational efficiency [6]. Also, in each down-sampling unit there is one conv\((1\times 1\times 1)\)-BN-ReLU which compresses the feature maps with a coefficient \(\theta \). A down-sampling unit is followed by one \(2\,\times \,2\,\times \,2\) max-pooling layer with a stride of \(2\times 2\times 2\). The up-sampling unit consists of one conv\((3\,\times \,3\,\times \,3)\)-BN-ReLU and one \(3\times 3\times 3\) transposed convolutional layer with a stride of \(2\,\times \,2\,\times \,2\). It has been shown that using bottleneck layers and compression aids in preventing overfitting [6].
Optimization is done by the Adam optimizer, with a constant learning rate of \(10^{-4}\). As the GTV is quite small in comparison with the background, the data is severely imbalanced. To tackle this issue [8], we employ the Dice similarity coefficient as a loss function of the network similar to [9]:
where \(s_i\in S\) is the binary segmentation of the GTV predicted by the network and \(g_i\in G\) is the ground truth segmentation.
3 Materials and Implementation
3.1 Dataset
This study includes two different datasets of chest CT scans. The first dataset was from 21 distinct patients who were treated for esophageal cancer between 2012 and 2014. For each patient, there were five repeat CT scans captured at different time points. For three time points, there was one 3D CT scan, and for two time points there were one 3D CT scan and one 4D CT scan, consisting of 10 breathing phases. The second dataset contains 29 distinct patients who were treated for esophageal cancer in 2016 and 2017, with a single 3D CT scan per patient. In both datasets, for each scan there was a corresponding esophageal GTV segmentation, delineated by a single experienced physician. Each volume contains \(58\text{- }108\) slices of \(512\times 512\) pixels and an average voxel size of \(0.98\times 0.98\times 3 \text { mm}^{3}\) which was resampled to a voxel size of \(1\times 1\times 3 \text { mm}^{3}\).
3.2 Augmentation and Training Details
We implemented DenseUnet in Google’s Tensorflow. The experiments are carried out using a GeForce GTX1080 Ti with 11 GB of GPU memory. For training the network, the dataset was divided into three distinct sets: 30 patients (390 volumes) for training, 6 patients (78 volumes) for validation, and 13 patients (85 volumes) for testing. In order to manage the GPU memory consumption and parallelize the patch selection during the training process, the patches were extracted randomly from the torso region, using a multi-threaded daemon process on the CPU and were fed to the network on the GPU. At the testing time the fully convolutional nature of the network is used, with zero padding to yield equal output size. The batch size is 20 and the number of training iterations is \(\sim \)10k. During every training iteration, the input patches were augmented by white noise extracted from a Gaussian distribution with zero mean and a random standard deviation between 0 and 5.
Table 1 summarizes the structure of the network. We used two different configurations of the proposed network, where we vary the number of the sub-blocks and also feature maps inside each sub-block.
4 Experiments and Results
Evaluation of the proposed method was done by the Dice Similarity Coefficient (\(\mathrm {DSC}\)), and mean surface distance (\(\mathrm {MSD}\)). As in some cases the networks additionally segment areas far away from the GTV, we also report the \(\mathrm {MSD}\) for the 95% best cases. In addition, precision and recall were reported.
The performance of DenseUnet with two configurations was compared with U-Net [10] and a modified version of U-Net dubbed DeepUnet\(_{122}\). The number of layers for the 3D U-Net network was 23 and the number of parameters 1.2M. The DeepUnet\(_{122}\) architecture is similar to DenseUnet\(_{122}\) but without the loop connections inside the dense blocks. In both U-Net and DeepUnet\(_{122}\), the Dice loss function was used instead of cross-entropy [10], to enable a fair comparison. Hence, the 3D U-Net in this paper is similar to V-Net [9].
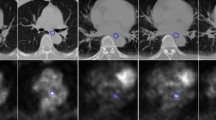
Figure 3(a–d) depicts the \(\mathrm {DSC}\), \(\mathrm {MSD}\) (mm), recall and precision for all networks on the test set. Figure 3(e) shows the cumulative frequency of the number of the scans for different \(\mathrm {DSC}\) values. As can be seen, for U-Net, 20 cases have a \(\mathrm {DSC} \le 0.5\), for DeepUnet\(_{122}\) still 15 cases are \(\le 0.5\), while this number is 10 and 8 for DenseUnet\(_{188}\) and DenseUnet\(_{122}\), respectively. A few cases have a low \(\mathrm {DSC}\) for all networks. A closer inspection revealed that in some cases there was a feeding tube, surgical clips or air pockets in the esophageal lumen present in the GTV. These cases were rarely seen in the training set. Figure 4 exemplifies the segmentation results for two normal GTVs, a GTV with a large air pocket in the lumen, and a GTV with a feeding tube present.
Example results, from left to right: original images; U-Net; DeepUnet\(_{122}\); DenseUnet\(_{188}\); DenseUnet\(_{122}\). The green contours depict the ground truth and the red overlays the network output. The 3\(^{\mathrm {rd}}\) and 4\(^\mathrm {th}\) rows show an example of the GTV containing an air cavity in the lumen and a feeding tube, respectively.
Table 2 yields a quantitative comparison of the segmentation performance for the networks. As can be seen, DenseUnet\(_{188}\) has the best value of MSD but DenseUnet\(_{122}\) has the best values in terms of \(\mathrm {DSC}\), 95% \(\mathrm {MSD}\) and median \(\mathrm {MSD}\). So, we propose DenseUnet\(_{122}\) as the final network.
5 Discussion and Conclusion
We proposed a 3D end-to-end fully convolutional CNN, called DenseUnet, for the segmentation of the esophageal GTV in CT images. DenseUnet leverages the ideas of dense blocks, in conjunction with down-sampling and up-sampling paths. This enables the network to extract contextual features deeply while retrieving image resolution and alleviating the problem of feature map explosion. We applied the proposed network to segment esophageal GTVs in 3D chest CT scans for the first time.
We trained and tested the proposed method on 553 chest CT scans from 49 distinct patients and achieved a \(\mathrm {DSC}\) value of \(0.73\pm 0.20\), and a 95% \(\mathrm {MSD}\) of \(3.07 \pm 1.86\) mm for the test scans, thereby outperforming U-Net. Eight (8/85) scans had a DSC \(<0.50\), mostly caused by the presence of air cavities and foreign bodies in the GTV, which was rarely seen in the training data.
To further enhance the robustness of the network we consider to increase the training data set (more foreign bodies) and use more elaborate data augmentation. Dilated convolutions may decrease the network size, and consequently make better use of the available training data as well. Combining with ROI-extraction techniques may lower the number of false positives.
In conclusion, the proposed network obtained promising results for the challenging problem of esophageal cancer segmentation on chest CT scans, comparing favorably to U-Net and earlier results found in the literature. The method therefore may assist the clinical workflow, especially when considering an online adaptive RT setting.
References
Thrift, A.P.: The epidemic of oesophageal carcinoma: where are we now? Cancer Epidemiol. 41, 88–95 (2016)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Fechter, T., Adebahr, S., Baltas, D., Ayed, I.B., Desrosiers, C., Dolz, J.: A 3D fully convolutional neural network and a random walker to segment the esophagus in CT. arXiv preprint arXiv:1704.06544 (2017)
Trullo, R., Petitjean, C., Nie, D., Shen, D., Ruan, S.: Fully automated esophagus segmentation with a hierarchical deep learning approach. In: IEEE ICSIPA, pp. 503–506 (2017)
Hao, Z., Liu, J., Liu, J.: Esophagus Tumor segmentation using fully convolutional neural network and graph cut. In: Jia, Y., Du, J., Zhang, W. (eds.) CISC 2017. LNEE, vol. 460, pp. 413–420. Springer, Singapore (2018). https://doi.org/10.1007/978-981-10-6499-9_39
Huang, G., Liu, Z., Weinberger, K.Q., van der Maaten, L.: Densely connected convolutional networks. In: IEEE CVPR, pp. 4700–4708 (2017)
Jégou, S., Drozdzal, M., Vazquez, D., Romero, A., Bengio, Y.: The one hundred layers tiramisu: fully convolutional densenets for semantic segmentation. In: IEEE CVPR Workshops, pp. 1175–1183 (2017)
Sudre, C.H., Li, W., Vercauteren, T., Ourselin, S., Jorge Cardoso, M.: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Cardoso, M.J., et al. (eds.) DLMIA/ML-CDS -2017. LNCS, vol. 10553, pp. 240–248. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67558-9_28
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: International Conference on 3D Vision, pp. 565–571 (2016)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Acknowledgements
Denis Shamonin is acknowledged for the torso extraction code.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Yousefi, S. et al. (2018). Esophageal Gross Tumor Volume Segmentation Using a 3D Convolutional Neural Network. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11073. Springer, Cham. https://doi.org/10.1007/978-3-030-00937-3_40
Download citation
DOI: https://doi.org/10.1007/978-3-030-00937-3_40
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00936-6
Online ISBN: 978-3-030-00937-3
eBook Packages: Computer ScienceComputer Science (R0)