Abstract
Segmentation is a key step for various medical image analysis tasks. Recently, deep neural networks could provide promising solutions for automatic image segmentation. The network training usually involves a large scale of training data with corresponding ground truth label maps. However, it is very challenging to obtain the ground-truth label maps due to the requirement of expertise knowledge and also intensive labor work. To address such challenges, we propose a novel semi-supervised deep learning framework, called “Attention based Semi-supervised Deep Networks” (ASDNet), to fulfill the segmentation tasks in an end-to-end fashion. Specifically, we propose a fully convolutional confidence network to adversarially train the segmentation network. Based on the confidence map from the confidence network, we then propose a region-attention based semi-supervised learning strategy to include the unlabeled data for training. Besides, sample attention mechanism is also explored to improve the network training. Experimental results on real clinical datasets show that our ASDNet can achieve state-of-the-art segmentation accuracy. Further analysis also indicates that our proposed network components contribute most to the improvement of performance.
D. Shen—This work was supported by the National Institutes of Health grant 1R01 CA140413.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Network Segmentation
- Ground Truth Label Maps
- Network Authority
- Ative Samples
- Semi-supervised Learning Scheme
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Recent development of deep learning has largely boosted the state-of-the-art segmentation methods [8, 11]. Among them, fully convolutional networks (FCN) [8], a variant of convolutional neural networks (CNN), is a recent popular choice for semantic image segmentation in both computer vision and medical image fields [8, 11, 13]. FCN trains neural networks in an end-to-end fashion by directly optimizing intermediate feature layers for segmentation, which makes it outperform the traditional methods that often regard the feature learning and segmentation as two separate tasks. UNet [11], an evolutionary variant of FCN, has achieved excellent performance by effectively combining high-level and low-level features in the network architecture. Generally, while being effective, the training of FCN (or UNet) requires a large amount of labeled data as there are millions of parameters in the network to be optimized. However, it is difficult to acquire a large training set with manually labeled ground-truth maps due to the following three factors: (a) manual annotation requires expertise knowledge; (b) it is time-consuming and tedious to annotate pixel-wise (voxel-wise) label maps; (c) it suffers from large intra- and inter-observer variability.
Several works have been done to address the aforementioned challenges [1, 2, 6]. To relieve the demand for large-scale labeled data, Bai et al. [1] proposed a semi-supervised deep learning framework for cardiac MR image segmentation, in which the segmented label maps from unlabeled data are incrementally included into the training set to refine the network. Baur et al. [2] introduced auxiliary manifold embedding in the latent space to FCN for semi-supervised learning in the MS lesion segmentation. In both cases, the unlabeled data information are fully involved in the model learning. However, certain regions of the unlabeled data may not be suitable for the learning due to their low-quality (automatically-) segmented label maps. To overcome such issues, we propose an attention based semi-supervised learning framework for medical image segmentation. Our framework is composed of two networks: (1) segmentation network and (2) confidence network. Specifically, we propose a fully convolutional adversarial learning scheme (i.e., using confidence network) to better train the segmentation network. The confidence map generated by the confidence network can provide us the trustworthy regions in the segmented label map from the segmentation network. Based on the confidence map, we further propose a region based semi-supervised loss to adaptively use part of unlabeled data for training the network. Since we can adopt unlabeled data to further train the segmentation network, the need of a large-scale training set can be alleviated accordingly. Our proposed algorithm has been applied to the task of pelvic organ segmentation, which is critical for guiding both biopsy and cancer radiation therapy. Experimental results indicate that our proposed algorithm can improve the segmentation accuracy, compared to other state-of-the-art methods. In addition, our proposed training strategies are also proved to be effective.
2 Method
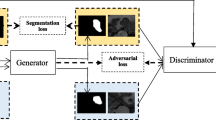
As mentioned above, the proposed ASDNet consists of two subnetworks, i.e., (1) segmentation network (denoted as S) and (2) confidence network (denoted as D). The architecture of our proposed framework is presented in Fig. 1.
To ease the description of the proposed algorithm, we first give the notations used throughout the paper. Given a labeled input image \(\mathbf {X} \in {R^{H \times W \times T}}\) with corresponding ground-truth label map \(\mathbf {Y} \in {Z^{H \times W \times T}}\), we encode it to one-hot format \(\mathbf {P} \in {R^{H \times W \times T \times C}}\), where \(C\) is the number of semantic categories in the dataset. The segmentation network outputs the class probability map \(\mathbf {\widehat{P}} \in {R^{H \times W \times T \times C}}\). Similarly, we regard an unlabeled image as \(\mathbf {U} \in {R^{H \times W \times T}}\). Therefore, the whole input image dataset can be defined by \(\mathbf {O} = \left\{ {\mathbf {X},\mathbf {U}} \right\} \).
2.1 Segmentation Network with Sample Attention
In ASDNet as shown in Fig. 1, the segmentation network can be any end-to-end segmentation network, such as FCN [8], UNet [11], VNet [9], and DSResUNet [13]. In this paper, we adopt a simplified VNet [9] (internal pool-conv-deconv layers are removed, and thus is denoted as SVNet) as the segmentation network to balance the performance and memory cost.
Multi-class Dice Loss: The class imbalance problem is usually serious in medical image segmentation tasks. To overcome it, we propose using a generalized multi-class Dice loss [12] as the segmentation loss, as defined below in Eq. 1:
where \(\mathbf {\pi }_c \) is the class balancing weight of category \(c\), \(\mathbf {{{\theta _S}}}\) is the parameters of segmentation network, and we set \({\pi _c} = 1/{\left( {\sum \limits _{h = 1}^H {\sum \limits _{w = 1}^W {\sum \limits _{t = 1}^T {{P_{h,w,t,c}}} } } } \right) ^2}\). \(\mathbf {\widehat{P}}\) is the predicted probability maps from the segmentation network: \(\mathbf {\widehat{P}} = S\left( {\mathbf {X},\mathbf {{\theta _s}}} \right) \).
Multi-class Dice Loss with Sample Attention: Besides the class imbalance problem, the network optimization also suffers from the issue of dominance by easy samples: the large number of easy samples will dominate network training, thus the difficult samples cannot be well considered. To address this issue, inspired by the focal loss [6] proposed to handle similar issue in detection networks, we propose a sample attention based mechanism to consider the importance of each sample during the training. The multi-class Dice loss with sample attention is thus defined below by Eq. 2:
where dsc is the average Dice similarity coefficient of the sample over different categories, e.g., different organ labels. Note that we re-compute the dsc in each iteration, but we don’t back-propagate gradient through it when training the networks. \(\beta \) is the sample attention parameter with a range of \(\left[ {0,5} \right] \). Following [6], we set \(\beta \) to 2 in this paper.
2.2 Confidence Network for Fully Convolutional Adversarial Learning
Adversarial learning is derived from the recent popular Generative Adversarial Network (GAN) [3]. It has achieved a great success in image generation and segmentation [3, 5, 10]. Hence, we also incorporate adversarial learning in our architecture to further improve the segmentation network. Instead of using CNN-based discriminator, we propose to use FCN-based discriminator to generate local confidence at local region.
Adversarial Loss of the Confidence Network: The training objective of the confidence network is the summation of binary cross-entropy loss over the image domain, as shown in Eq. 3. Here, we use S and D to denote the segmentation and confidence networks, respectively.
where
where \({\mathbf {X}}\) and \({\mathbf {P}}\) represent the input data and its corresponding manual label map (one-hot encoding format), respectively. \({\mathbf {\theta _d}}\) is network parameters for the confidence network.
Adversarial Loss of the Segmentation Network: For segmentation network, besides the multi-class Dice loss with sample attention as defined in Eq. 2, there is another loss from D working as “variational” loss. It enforces higher-order consistency between ground-truth segmentation and automatic segmentation. In particular, the adversarial loss (“ADV”) to improve S and fool D can be defined by Eq. 5.
2.3 Region-Attention Based Semi-supervised Learning
Since our discriminator (i.e., confidence network) could provide local confidence information over the image domain, we use such information in the semi-supervised setting to include unlabeled data for improving segmentation accuracy, and the similar strategy has been explored in [5].
Specifically, given an unlabeled image \( \mathbf {U} \), the segmentation network will first produce the probability map \(\mathbf {\widehat{P}} = S\left( \mathbf {U} \right) \), which will be then used by the trained confidence network to generate a confidence map \(\mathbf {M} = D( {\mathbf {\widehat{P}}} )\), indicating where the confident regions of the prediction results are close enough to the ground truth label distribution. The confident regions can be easily obtained by setting a threshold (i.e., \(\gamma \)) to the confidence map. In this way, we can use these confident regions as masks to select parts of unlabeled data and their segmentation results to enrich the set of supervised training data. Thus, our proposed semi-supervised loss can be defined by Eq. 6.
where \(\mathbf {{\overline{P} }}\) is the one-hot encoding of \(\mathbf {\widehat{Y}}\), and \(\mathbf {\widehat{Y}} = \arg \max ( \mathbf {{\widehat{P}}} )\). [] is the indicator function. Similar to \(dsc\) in Eq. 2, \(\mathbf {\overline{P}} \) and the value of indicator function are re-computed in each iteration.
Total Loss for Segmentation Network: By summing the above losses, the total loss to train the segmentation network can be defined by Eq. 7.
where \({\lambda _1}\) and \({\lambda _2}\) are the scaling factors to balance the losses. They are selected at 0.03 and 0.3 after trails, respectively.
2.4 Implementation Details
PytorchFootnote 1 is adopted to implement our proposed ASDNet shown in Fig. 1. We adopt Adam algorithm to optimize the network. The input size of the segmentation network is \(64 \times 64 \times 16\). The network weights are initialized by the Xavier algorithm, and weight decay is set to be 1e–4. For the network biases, we initialize them to 0. The learning rates for the segmentation and confidence network are initialized to 1e–3 and 1e–4, followed by decreasing the learning rate 10 times every 3 epochs. Four Titan X GPUs are utilized to train the networks.
3 Experiments and Results
Our pelvic dataset consists of 50 prostate cancer patients from a cancer hospital, each with one T2-weighted MR image and corresponding manually-annotated label map by medical experts. In particular, the prostate, bladder and rectum in all these MRI scans have been manually segmented, which serve as the ground truth for evaluating our segmentation method. Besides, we have also acquired 20 MR images from additional 20 patients, without manually-annotated label maps. All these images were acquired with 3T MRI scanners. The image size is mostly \(256\times 256\times \left( {120{-}176} \right) \), and the voxel size is \(1\times 1\times 1~\text {mm}^3\).
Five-fold cross validation is used to evaluate our method. Specifically, in each fold of cross validation, we randomly chose 35 subjects as training set, 5 subjects as validation set, and the remaining 10 subjects as testing set. We use sliding windows to go through the whole MRI for prediction for a testing subject. Unless explicitly mentioned, all the reported performance by default is evaluated on the testing set. As for evaluation metrics, we utilize Dice Similarity Coefficient (DSC) and Average Surface Distance (ASD) to measure the agreement between the manually and automatically segmented label maps.
3.1 Comparison with State-of-the-art Methods
To demonstrate the advantage of our proposed method, we also compare our method with other five widely-used methods on the same dataset as shown in Table 1: (1) multi-atlas label fusion (MALF), (2) SSAE [4], (3) UNet [11], (4) VNet [9], and (5) DSResUNet [13]. Also, we present the performance of our proposed ASDNet.
Table 1 quantitatively compares our method with the five state-of-the-art segmentation methods. We can see that our method achieves better accuracy than the five state-of-the-art methods in terms of both DSC and ASD. The VNet works well in segmenting bladder and prostate, but it cannot work very well for rectum (which is often more challenging to segment due to the long and narrow shape). Compared to UNet, DSResUNet improves the accuracy by a large margin, indicating that residual learning and deep supervision bring performance gain, and thus it might be a good future direction for us to further improve our proposed method. We also visualize some typical segmentation results in Fig. 2, which further show the superiority of our proposed method.
3.2 Impact of Each Proposed Component
As our proposed method consists of several designed components, we conduct empirical studies below to analyze them.
Impact of Sample Attention: As mentioned in Sect. 2.1, we propose a sample attention mechanism to assign different importance for different samples so that the network can concentrate on hard-to-segment examples and thus avoid dominance by easy-to-segment samples. The effectiveness of sample attention mechanism (i.e., AttSVNet) is further confirmed by the improved performance, e.g., 0.82%, 1.60% and 1.81% DSC performance improvements (as shown in Table 2) for bladder, prostate and rectum, respectively.
Impact of Fully Convolutional Adversarial Learning: We conduct more experiments for comparing with the following three networks: (1) only segmentation network; (2) segmentation network with a CNN-based discriminator [3]; (3) segmentation network with a FCN-based discriminator (i.e., confidence network). Performance in the middle of Table 2 indicates that adversarial learning contributes a little bit to improving the results as it provides a regularization to prevent overfitting. Compared with CNN-based adversarial learning, our proposed FCN-based adversarial learning further improves the performances by 0.90% in average. This demonstrates that fully convolutional adversarial learning works better than the typical adversarial learning with a CNN-based discriminator, which means the FCN-based adversarial learning can better learn structural information from the distribution of ground-truth label map.
Impact of Semi-supervised Loss: We apply the semi-supervised learning strategy with our proposed ASDNet on 50 labeled MRI and 20 extra unlabeled MRI. The comparison methods are semiFCN [1] and semiEmbedFCN [2]. We use the AttSVNet as the basic architecture of these two methods for fair comparison. The evaluation of the comparison experiments are all based on the labeled dataset, and the unlabeled data involves only in the learning phase. The experimental results in Table 2 show that our proposed semi-supervised strategy works better than the semiFCN and the semiEmbedFCN. Moreover, it is worth noting that the adversarial learning on the labeled data is important to our proposed semi-supervised scheme. If the segmentation network does not seek to fool the discriminator (i.e., AttSVNet+Semi), the confidence maps generated by the confidence network would not be meaningful.
3.3 Validation on Another Dataset
To show the generalization ability of our proposed algorithm, we conduct additional experiments on the PROMISE12-challenge dataset [7]. This dataset contains 50 subjects, each with a pair of MRI and its manual label map (where only prostate was annotated). Five-fold cross validation is performed to evaluate the performance of all comparison methods. Our proposed algorithm again achieves very good performance in segmenting prostate (i.e., 0.900 in terms of DSC), and it is also very competitive compared to the state-of-the-art methods applied to this dataset in the literature [9, 13]. These experimental results indicate a good generalization capability of our proposed ASDNet.
4 Conclusions
In this paper, we have presented a novel attention-based semi-supervised deep network (ASDNet) to segment medical images. Specifically, the semi-supervised learning strategy is implemented by fully convolutional adversarial learning, and also region-attention based semi-supervised loss is adopted to effectively address the insufficient data problem for training the complex networks. By integrating these components into the framework, our proposed ASDNet has achieved significant improvement in terms of both accuracy and robustness.
References
Bai, W., et al.: Semi-supervised learning for network-based cardiac MR image segmentation. In: Descoteaux, M., et al. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 253–260. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_29
Baur, C., Albarqouni, S., Navab, N.: Semi-supervised deep learning for fully convolutional networks. In: Descoteaux, M. (ed.) MICCAI 2017. LNCS, vol. 10435, pp. 311–319. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_36
Goodfellow, I., et al.: Generative adversarial nets. In: NIPS (2014)
Guo, Y., et al.: Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE TMI 35, 1077–1089 (2016)
Hung, W.-C., et al.: Adversarial learning for semi-supervised semantic segmentation. arXiv preprint arXiv:1802.07934 (2018)
Lin, T.-Y., et al.: Focal loss for dense object detection. arXiv preprint arXiv:1708.02002 (2017)
Litjens, G.: Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. MedIA 18(2), 359–373 (2014)
Long, J., et al.: Fully convolutional networks for semantic segmentation. In: CVPR, pp. 3431–3440 (2015)
Milletari, F., et al.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 3DV, pp. 565–571. IEEE (2016)
Nie, D., et al.: Medical image synthesis with context-aware generative adversarial networks. In: Descoteaux, M., et al. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 417–425. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_48
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sudre, C.H., et al.: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. DLMIA/ML-CDS -2017. LNCS, vol. 10553, pp. 240–248. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67558-9_28
Yu, L., et al.: Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images. In: AAAI (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Nie, D., Gao, Y., Wang, L., Shen, D. (2018). ASDNet: Attention Based Semi-supervised Deep Networks for Medical Image Segmentation. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11073. Springer, Cham. https://doi.org/10.1007/978-3-030-00937-3_43
Download citation
DOI: https://doi.org/10.1007/978-3-030-00937-3_43
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00936-6
Online ISBN: 978-3-030-00937-3
eBook Packages: Computer ScienceComputer Science (R0)