Abstract
The presence and severity of emphysema, scored visually on computed tomography (CT) using a classification system developed by the Fleischner Society, is a clinically significant index of disease severity. Since visual assessment can be subjective and is time consuming, our purpose was to evaluate the potential of a deep learning method for automatic grading of emphysema. The study cohort included 8213 subjects enrolled in the COPDGene study. Baseline CT and visual scores on 2500 subjects were used to train a deep learning model for classification of centrilobular emphysema according to the Fleischner system. The model was then used to predict emphysema scores on 5713 subjects not included in the training set. Predictions were compared with visual emphysema scores, pulmonary function tests (PFTs), smoking history and St. George Respiratory Questionnaire (SGRQ). Agreement between visual emphysema scores and those generated automatically was moderate (weighted \(\kappa \) = 0.60, p < 0.0001). Emphysema scores predicted by the deep learning model showed significant associations with PFTs, smoking history and SGRQ, similar to those seen in comparison with visual scores.
This work was supported by grants R01HL089897 and R01HL089856 from the National Heart, Lung, and Blood Institute. The Genetic Epidemiology of COPD (COPDGene) project is also supported by the COPD Foundation through contributions made to an industry advisory board representing AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, Sunovion, and GlaxoSmithKline.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Chronic obstructive pulmonary disease (COPD), the third leading cause of death in the U.S., is a heterogeneous group of lung disorders, including a range of patterns of emphysema, chronic bronchitis, and non-emphysematous obstruction due to small-airway disease [7]. CT plays an important role in assessment of COPD. While quantitative image analysis techniques such as lung densitometry are able to detect and quantify emphysema [13], visual assessment has remained necessary for complete characterization of emphysema patterns [7].
To facilitate data comparison in research and clinical trials, and to improve diagnostic accuracy, the Fleischner Society developed a structured system for visual classification of phenotypic abnormalities on CT in subjects with COPD [6]. The system provides a six point ordinal scale (absent, trace, mild, moderate, confluent, and advanced destructive) for grading the severity of centrilobular emphysema, the prototypical form of emphysema seen in cigarette smokers. This approach was applied in the COPDGene study where baseline CT scans were retrospectively visually scored by trained analysts. Visual scores for presence and severity of centrilobular emphysema were found to be significantly associated with genetic loci previously associated with COPD [5] and with increased mortality risk [7]. Visual assessment required substantial effort, however, with multiple analysts working nearly four years to score approximately ten thousand scans.
The purpose of this study was to assess the feasibility of a deep learning model for automatic emphysema classification on CT. We hypothesized that the Fleischner system, a structured scale developed by domain experts, would provide an effective supervisory signal for training. To develop a model capable of predicting subject-level scores from volumetric CT, which are difficult to process at full resolution due to memory constraints of graphics processing units (GPUs), we combined a convolutional neural network (CNN) architecture with a long short-term memory (LSTM) layer. LSTMs are a type of recurrent neural network capable of learning dependencies in sequence data. They have been used with some success to classify data such as frame sequences from video clips [1]. This enabled efficient prediction using features extracted from axial image sequences sampled over a lung volume. We tested the approach by comparing predictions with visual scores and clinical parameters including pulmonary function tests and Global initiative for Obstructive Lung Disease (GOLD) stage.
2 Methods
2.1 Study Population
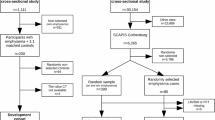
COPDGene is a prospective and multicenter investigation focused on the genetic epidemiology of COPD (ClinicalTrials. gov: NCT00608764). All subjects underwent volumetric inspiratory and expiratory CT using a standardized protocol [9]. Inspiratory CT on 8213 subjects were included in this work. The cohort was partitioned so that scans and visual scores on 2500 subjects were used for training and data on the remaining 5713 subjects were reserved for testing. Distribution of emphysema scores in training data was n = 774, 435, 473, 431, 275, 112 for Fleischner scores 0, 1, 2, 3, 4, and 5, respectively. Additional variables including GOLD stage, Forced Expiratory Volume in the first second percent predicted (FEV1\({\%}\)pred), FEV1/Forced Vital Capacity ratio (FEV1/FVC), St. George Respiratory Questionnaire (SGRQ) and smoking history (pack-years) were retrieved from the study database for the test partition. The SGRQ is a respiratory health-related quality of life questionnaire where higher scores correspond to greater impairment [15]. Quantitative CT emphysema score calculated as the percentage of lung voxels with intensity less than −950 Hounsfield Units (LAA-950) was also used for comparison in test subjects.
2.2 Visual Scoring
Each CT study had been retrospectively visually scored by two trained analysts using the scale 0 = absent, 1 = trace, 2 = mild, 3 = moderate, 4 = confluent, and 5 = advanced destructive [7]. Analyst agreement was assessed periodically throughout the study as good to excellent (weighted \(\kappa \) range 0.71–0.80). Analyst discordances larger than one point in score were adjudicated by a thoracic radiologist. Mean analyst score rounded down to the nearest integer was recorded as the final value.
2.3 Classification Algorithm
In an initial process, segmentation was performed on all CTs using an automatic lung segmentation function included in an open source library [11, 12]. Using Python and PyTorch [8] we implemented a combined CNN-LSTM network architecture [1] designed to predict visual centrilobular emphysema score from a sequence of 25 axial images. Axial images were sampled at evenly-spaced intervals over the height of each lung segmentation volume, excluding the upper and lowermost 5 mm. Figure 1 represents the architecture of the CNN-LSTM model. Briefly, the components of the network include four blocks consisting of two dimensional (2D) convolutions, rectified linear unit (RELU) activation and max pooling. The four 2D convolutional layers have 32 6\(\,\times \,\)6, 96 3\(\,\times \,\)3, 256 3\(\,\times \,\)3 and 384 3\(\,\times \,\)3 filters, respectively. The first two max pooling layers have stride 3 and the second two max pooling layers have stride 4. The input images are fed through the CNN portion separately to extract features, which are concatenated into a sequence and passed to the LSTM layer followed by a dense layer before output. The loss function is negative log likelihood.
The model was trained using CT scans and visual centrilobular emphysema scores, expressed as integers 0–5, on 2500 subjects. Some data augmentation was used, including in-plane image translations and offsets in sampling of axial images.
2.4 Statistical Analysis
Weighted \(\kappa \) statistics between analysts visual scores and automatic scores were computed. Descriptive statistics between CNN-LSTM emphysema scores and demographic and clinical parameters were computed. One-way analysis of variance (ANOVA) was used to test for significant differences in FEV1\({\%}\)pred., FEV1/FVC, SGRQ, LAA-950 and smoking history values stratified by emphysema severity scores. Chi-square tests of independence were used to compare centrilobular emphysema scores with GOLD stage. Statistical calculations were performed using R version 3.4.4 (2018-03-15). A p-value of <0.05 was considered statistically significant.
3 Results
Agreement between visual emphysema scores and those generated automatically by the CNN-LSTM was moderate (weighted \(\kappa \) = 0.60, p < 0.0001). Table 1 shows a confusion matrix comparing visual and automatic emphysema scores.
Table 2 shows comparison between visual emphysema score and clinical variables. ANOVA showed that more severe visual emphysema scores were significantly associated with diminished FEV1\(\%\)pred and FEV1/FVC as well as increased SGRQ, LAA-950 and smoking pack-years (p < 0.0001). Chi-square test of independence examining the relationship between visual score and GOLD stage was significant, \(\chi ^2\) (df = 25, n = 5713) = 2716.4, p\(\,<\,\)0.0001. Similarly, Table 3 shows comparison between CNN-LSTM emphysema scores and clinical variables. ANOVA shows that more severe emphysema grades predicted by the CNN-LSTM were associated with more severe clinical measures. Chi-square test of independence examining the relationship between CNN-LTSM score and GOLD stage was also significant, \(\chi ^2\) (df = 25, n = 5713) = 3203.4, p < 0.0001.
4 Discussion
The Fleischner Society visual scoring system is a clinically significant index of COPD severity that is associated with mortality risk. We have developed a CNN-LSTM model that is capable of automatic classification of centrilobular emphysema pattern on CT according to the Fleischner scale. In a group of 5713 test subjects, automatically scored emphysema patterns showed moderate agreement with visual scores and significant associations with GOLD stage, pulmonary function, SGRQ, LAA-950 and smoking pack-years. While agreement between visual and automatic emphysema scores in this test set is somewhat modest, similar statistical associations are seen when comparing visual and CNN-LSTM emphysema scores with other clinical parameters, suggesting that the CNN-LSTM learns to classify clinically significant features.
Deep learning has become the dominant approach in medical image analysis [16] for applications ranging from segmentation [10] and detection [14] to diagnosis [2]. One challenge when designing CNN architectures is to manage the total number of model parameters considering memory constraints of available GPUs. This is generally the case in applications using volumetric chest CT. Other researchers have shown that limiting model input to a small number (1–4) of relevant “canonical” slices, selected in pre-processing using anatomy detection methods, can be effective [3, 4]. In the present study we instead used an LSTM architecture, which made it possible to limit the number of convolutional weights while still using 25 full-resolution axial images as input.
This study has some limitations. COPDGene uses a well-defined CT protocol and study images are very consistent. Performing both training and testing on images from this cohort may produce optimistic results. We also did not perform systematic comparisons of different CNN architectures. Future work will incorporate mortality and longitudinal assessment for further clinical validation.
5 Conclusions
A combined CNN-LSTM architecture provides an efficient model for subject-level prediction and can be trained to perform automatic classification of centrilobular emphysema on CT.
References
Donahue, J., et al.: Long-term recurrent convolutional networks for visual recognition and description. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2625–2634 (2015)
Esteva, A., et al.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115 (2017)
González, G., et al.: Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am. J. Respir. Crit. Care Med. 197(2), 193–203 (2018)
González, G., Washko, G.R., San José Estépar, R.a.: Deep learning for biomarker regression: application to osteoporosis and emphysema on chest CT scans. In: SPIE Medical Imaging, vol. 10574 (2018)
Halper-Stromberg, E., et al.: Visual assessment of chest computed tomographic images is independently useful for genetic association analysis in studies of chronic obstructive pulmonary disease. Ann. Am. Thorac. Soc. 14(1), 33–40 (2017)
Lynch, D.A., et al.: CT-definable subtypes of chronic obstructive pulmonary disease: a statement of the fleischner society. Radiology 277(1), 192–205 (2015)
Lynch, D.A.: CT-based visual classification of emphysema: association with mortality in the COPDGene study. Radiology 288, 859–866 (2018)
Paszke, A., et al.: Automatic Differentiation in PyTorch (2017)
Regan, E.A., et al.: Genetic epidemiology of COPD (COPDGene) study design. COPD J. Chronic Obstructive Pulm. Dis. 7(1), 32–43 (2011)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Ross, J.C., et al.: Lung extraction, lobe segmentation and hierarchical region assessment for quantitative analysis on high resolution computed tomography images. In: Yang, G.-Z., Hawkes, D., Rueckert, D., Noble, A., Taylor, C. (eds.) MICCAI 2009. LNCS, vol. 5762, pp. 690–698. Springer, Heidelberg (2009). https://doi.org/10.1007/978-3-642-04271-3_84
Ross, J., Harmouche, R., Onieva, J., Diaz, A., Washko, G., Estepar, R.S.J.: Chest imaging platform: an open-source library and workstation for quantitative chest imaging. Am. J. Respir. Crit. Care Med. 191, A4975 (2015)
Schroeder, J.D., et al.: Relationships between airflow obstruction and quantitative CT measurements of emphysema, air trapping, and airways in subjects with and without chronic obstructive pulmonary disease. Am. J. Roentgenol. 201(3), W460–W470 (2013)
Setio, A.A.A., et al.: Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 35(5), 1160–1169 (2016)
Vestbo, J.: Evaluation of COPD longitudinally to identify predictive surrogate endpoints (ECLIPSE). Eur. Respir. J. 31, 869–873 (2008)
Zhou, S.K., Greenspan, H., Shen, D.: Deep Learning for Medical Image Analysis. Academic Press, San Diego (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Humphries, S.M., Notary, A.M., Centeno, J.P., Lynch, D.A. (2018). Automatic Classification of Centrilobular Emphysema on CT Using Deep Learning: Comparison with Visual Scoring. In: Stoyanov, D., et al. Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO BIA TIA 2018 2018 2018. Lecture Notes in Computer Science(), vol 11040. Springer, Cham. https://doi.org/10.1007/978-3-030-00946-5_32
Download citation
DOI: https://doi.org/10.1007/978-3-030-00946-5_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00945-8
Online ISBN: 978-3-030-00946-5
eBook Packages: Computer ScienceComputer Science (R0)