Abstract
Neural networks have emerged as a successful tool to solve end-to-end classification problems, potentially applicable in many diagnostic settings once trained with a sufficient number of existing annotations. Nevertheless, in such training it is often nontrivial to enter already available domain knowledge. We herein propose a simple approach of inputing any such information as additional layers to a network. This may then yield better performance by allowing for networks with fewer parameters that can be tuned with fewer annotations and with better generalization capabilities. This can also allow for interpretability of a deep network, by quantifying attribution to such additional inputs. We study this approach for the task of skin lesion classification, where we focus on prior knowledge in the form of pigment networks as they are known visual indicators of certain skin lesions, e.g. melanoma. We used a public dataset of dermoscopic images, where a low number of feature segmentations and a high number of classifications are provided in disjoint datasets. By including information from learned pigment network segmentations, the recall for malignant melanoma was seen to increase from 0.213 to 0.4. To help interpret the results, we also quantified the “attention” to pigment networks paid by the deep classifier both location- and channel-wise.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Skin cancer is one of the most prevalent types of cancer [1, 2] and there is a growing need for accurate and scalable decision support systems for skin diseases. To assist doctors in making correct diagnoses, decision support systems can be trained on dermoscopic images, the same type of input data that dermatologists often use for an initial assessment.
The International Skin Imaging Collaboration (ISIC) [7, 10] provides public datasets of dermoscopic images and organizes challenges where state-of-the-art (SoA) methods in this field can compete. These datasets allow researchers to design data-driven systems for the detection of skin diseases. Although in recent years high accuracy has been achieved with different Deep Learning approaches [7, 9, 10], most methods do not provide a mechanism to make use of prior medical knowledge.
In this work, we present a novel approach that tackles this issue. We aim at leveraging the predictive power of a deep convolutional neural network (CNN) while providing functionalities to understand which factors influence the network’s prediction. We further quantify the attention that the trained classifier pays to each feature channel and image location, as a means to demonstrate our conclusions.
Annotation of dermascopic structures overlaid on images [7].
Related Work. Esteva et al. recently presented a CNN-based approach that outperformed certified dermatologists at differentiating benign and malignant lesions [9]. They used transfer learning and a disease partitioning algorithm for the generation of optimal training classes. They further computed saliency maps that highlight the importance of every pixel for the final prediction. However, the saliency maps provide only little interpretable information, such as the fact that the network mainly focuses on pixels belonging to the lesion rather than on the background.
Codella et al. used a mixture of hand-coded features and features extracted by deep CNNs to achieve SoA results on the dataset of the ISBI 2016 “Skin Lesion Analysis Towards Melanoma Detection” challenge [6, 10]. Despite leveraging color features and shape descriptors for lesions, their approach does not facilitate an intuitive way of understanding the system’s predictions.
The extensive work of López-Labraca et al. [13] is closely related to our approach. They employed sophisticated, hand-crafted filters to detect relevant dermoscopic structures (see Fig. 1). For a given lesion image, malignancy scores of different dermoscopic structures were computed and then combined to form a single diagnosis (malignant or benign). The authors were able to generate comprehensive reports containing the final diagnosis and the detected structures along with their respective malignancy scores. Nonetheless, their proposed approach requires extensive feature engineering and, in contrast to deep learning methods, is limited to features that are already known to dermatologists.
Building on the same idea, González-Díaz presented a method that used dermoscopic structures in combination with ResNet50, a deep residual network [8, 11]. An input image was fed into a segmentation network that produced probability maps of eight different dermoscopic structures. These maps were then used to modulate the latent representation of the input image at a hidden layer in the ResNet50. Using this CNN-based method, González-Díaz achieved the best score for the detection of seborrheic keratosis in the ISBI 2017 “Skin Lesion Analysis Towards Melanoma Detection” challenge [7]. However, despite making use of known dermoscopic structures, this method does not provide interpretable information as the work of López-Labraca et al., where hand-crafted features were employed [13]. Furthermore, it is unclear to what extent the segmentations of the dermoscopic structures influence the final diagnosis.
2 Methods
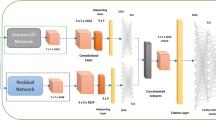
Overview. Our method consists of two stages. Given an input image of a skin lesion, we first employ a segmentation network (SN) to detect dermoscopic structures that dermatologists consider to be important for disease classification (see Fig. 1). We focus on pigment networks as they are known indicators for malignant melanoma and benign nevi. Furthermore, they are the dermoscopic structures that are segmented with the highest confidence by our SN. In a second stage, the output of the network is stacked on top of the existing RGB channels of the original image, and then the resulting four-channel input is used to train a classifier network (CN) for each considered type of disease.
Additionally, we introduce two measures of attention given by the classifier network, namely the channel-wise and location-wise attention. These measures allow us to quantify how much attention the classifier is paying to the provided dermoscopic structure compared to the rest of the input data.
Material and Dataset. For the detection of pigment networks, we trained our SN with the dataset provided by the ISBI 2017 “Skin Lesion Analysis Towards Melanoma Detection” challenge [7]. It consists of 2000 training images and 600 testing images with superpixel level annotations of different types of dermoscopic structures, one of which is the pigment network. Every image is labeled with one of three classes: Melanoma, nevus, or seborrheic keratosis.
For the disease classification task, we trained our CN on the datasets released for the “ISIC 2018: Skin Lesion Analysis Towards Melanoma Detection” challenge [7, 17]. It comprises 10,015 images belonging to one of seven classes: melanoma, melanocytic nevus, basal cell carcinoma, actinic keratosis, benign keratosis, dermatofibroma, or vascular lesion. For simplicity, we will from now on call the datasets DS2017 and DS2018, respectively.
Methodology Overview. Since the images exhibit varying dimensions, they were resized to \(224\times 224\) pixels. Both datasets were augmented with random rotations of \(90^\circ \), \(180^\circ \) and \(270^\circ \), as well as vertical and horizontal flips. The algorithms for pigment network segmentation SN and disease classification CN were implemented in Tensorflow [3], both by extending the code provided by [5].
Detection of Pigment Networks. The detection task was formulated as a pixel-wise binary segmentation problem with a foreground and a background class. Due to the large class imbalance by background pixels, we reduced the original training set to a subset containing only those images where a pigment network was annotated. Note that this may result in a biased segmenter SN because it has been trained to always detect a pigment network somewhere in the image. Nevertheless, the subsequent disease classifier CN still sees the actual image and may choose to ignore this segmented area, if that does not facilitate the classification. Accordingly, our motivation was to have an (over-)sensitive SN, in order to let the subsequent CN decide how much importance to give to the allegedly-detected pigment network. Although such a two-step approach can be argued to potentially be inferior to an end-to-end solution, the former allows us to facilitate dedicated datasets and train targeted models, giving us more control over each step.
For SN, we employed a shallow U-Net [14] that outputs probability maps for the occurrence of pigment networks. To further alleviate the problem of background dominance, the Sørensen-Dice coefficient was used as loss function.
Disease Classification. We evaluated two types of classifiers: (i) ResNet50 (with 50 layers) pre-trained with images from the 2014 ImageNet Large Scale Visual Recognition Challenge [15], and (ii) the shallower ResNet18 (with 18 layers) proposed by He et al. [11], which we trained from scratch.
Attention. Our attention measures are based on so-called attribution methods. Given a deep CNN with input \(x = [x_1, ... , x_N] \in \mathbb {R}^N\) and output \(f(x) = [f_1(x), ... , f_C(x)] \in \mathbb {R}^C\), attribution methods compute the contribution \(R_{i,c}\) of every input pixel \(x_i\) to a specific target neuron \(f_c\). Different types of attribution methods have been proposed in the past, such as the perturbation- and gradient-based approaches [4]. We herein employed a simple method of input \(\times \) partial-derivative [16], which is fast and worked for generating successful attribution maps for our purposes (see Fig. 2). This metric defines attribution component \(R_{i,c}\) of an input pixel \(x_i\) to a target neuron \(f_c\) as follows:

Based on (1) we propose two quantities to measure the attention that a CNN pays to each input channel and image location.
Channel-wise attention (\(A_c\)) is defined as the ratio of contribution from a particular structure channel c to the contributions of all K channels:

Location-wise attention (\(A_L\)) captures the local attention in the image space, again as a ratio. The numerator contains the contributions of all channels except for the dermoscopic structure channel c. The contributions are weighted with p, the local probability of the dermoscopic structure; and the denominator contains the corresponding unweighted contributions:

We used the implementation of Ancona et al.Footnote 1 to compute the contribution values and to generate the attribution maps.
3 Results and Discussion
Segmentation. Examples for segmentations of pigment networks are depicted in Fig. 3. We trained and tested on DS2017 because at the moment of this writing, SoA results for DS2018 are not yet publicly available for comparison. As seen in Table 1, our pigment network segmentation results are not as accurate but comparable to the SoA results by Kawahara & Hamarneh from the ISBI 2017 “Skin Lesion Analysis Towards Melanoma Detection” challenge [7] in dermoscopic structure segmentation [12]. Note that our goal herein was not to perfect the SN stage, but rather to investigate if and how any information that SN provides can be further used in classification.
Classification. For the classification experiments, we used the DS2018 training set and applied an 80%-10%-10%-split into training, validation and test set. Table 2 and 3 show the classification scores of ResNet18 and ResNet50 with and without additional pigment network channel.
For ResNet18, the ROC AUC values do not change significantly when the pigment network channel is added. Despite a slight overall decrease in ROC AUC values, Table 3 shows that the F1-score for the crucial melanoma class increases thanks to a clear improvement of the recall from 0.213 to 0.400. This is even better than the recall obtained by the much more complex ResNet50. In terms of ROC AUC values, ResNet50 still performs best. Notice however, that adding the pigment network channel to the input of ResNet50 actually leads to lower ROC AUC values, recall, and F1-scores.
As seen in Table 4, the channel-wise attention \(A_c\) as well as the location-wise attention \(A_l\) for melanoma and melanocytic nevus are clearly higher in the case of ResNet18. This suggests that ResNet50 is not focusing its attention on the parts of the image that are medically relevant. The pre-trained ResNet50 may require more sophisticated fine-tuning if an additional channel is to be used. In our approach, only the weights of the first convolutional layer and the final fully-connected layer were learned whereas all weights in-between were pre-trained and frozen. Since the images from the 2014 ImageNet Large Scale Visual Recognition Challenge [15] are very different from dermoscopic images, it might be beneficial to use more general pre-trained feature representations from a higher layer and start learning from there. However, this is in turn computationally more expensive.
4 Conclusion
We showed that the recall and the F1-score for the detection of melanoma can be improved by providing a CNN with an additional input channel that contains relevant prior knowledge. Furthermore, we demonstrated that our proposed attention measures can help to identify where a CNN focuses its attention. In a next step, one might consider integrating more than just information about pigment networks in the input. Other dermoscopic structures such as streaks and dots could be used to further improve existing classifiers, e.g. for non-melanocytic lesions.
Notes
- 1.
DeepExplain repository 24 May 2018: https://github.com/marcoancona/DeepExplain.
References
Cancer in Australia 2017. Technical Report 101, Australian Institute of Health and Welfare. AIHW, Canberra, February 2017
U.S. cancer statistics working group. U.S. cancer statistics data visualizations tool. Technical report, Centers for Disease Control and Prevention and National Cancer Institute, June 2018. www.cdc.gov/cancer/dataviz
Abadi, M., et al.: TensorFlow: large-scale machine learning on heterogeneous systems (2015). tensorflow.org
Ancona, M., Ceolini, E., Oztireli, C., Gross, M.: Towards better understanding of gradient-based attribution methods for deep neural networks. In: 6th International Conference on Learning Representations (ICLR 2018) (2018)
Baumgartner, C.F., Koch, L.M., Pollefeys, M., Konukoglu, E.: An exploration of 2D and 3D deep learning techniques for cardiac MR image segmentation. In: Pop, M., et al. (eds.) STACOM 2017. LNCS, vol. 10663, pp. 111–119. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75541-0_12
Codella, N.C.F., et al.: Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J. Res. Dev. 61(4/5), 5:1–5:15 (2017)
Codella, N.C.F., et al.: Skin lesion analysis toward melanoma detection: a challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), hosted by the International Skin Imaging Collaboration (ISIC). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 168–172. IEEE, April 2018
Diaz, I.G.: DermaKNet: Incorporating the knowledge of dermatologists to convolutional neural networks for skin lesion diagnosis. IEEE J. Biomed. Health Inf., 1 (2018)
Esteva, A., Kuprel, B., Novoa, R.A., Ko, J., Swetter, S.M., Blau, H.M., Thrun, S.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115–118 (2017)
Gutman, D., et al.: Skin lesion analysis toward melanoma detection: A challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). ArXiv e-prints, May 2016
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778, June 2016
Kawahara, J., Hamarneh, G.: Fully convolutional neural networks to detect clinical dermoscopic features. IEEE J. Biomed. Health Inf., 1 (2018)
López-Labraca, J., Fernández-Torres, M.Á., González-Díaz, I., Díaz-de María, F., Pizarro, Á.: Enriched dermoscopic-structure-based CAD system for melanoma diagnosis. Multimedia Tools Appl. 77(10), 12171–12202 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Russakovsky, O., et al.: ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 115(3), 211–252 (2015)
Shrikumar, A., Greenside, P., Shcherbina, A., Kundaje, A.: Not just a black box: learning important features through propagating activation differences. arXiv preprint arXiv:1605.01713 (2016)
Tschandl, P., Rosendahl, C., Kittler, H.: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 5, 180161 (2018). https://doi.org/10.1038/sdata.2018.161
Acknowledgments
Support was provided by IBM Research Zurich, Switzerland and the Promedica Foundation, Chur, Switzerland.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Thandiackal, K., Goksel, O. (2018). A Structure-Aware Convolutional Neural Network for Skin Lesion Classification. In: Stoyanov, D., et al. OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis. CARE CLIP OR 2.0 ISIC 2018 2018 2018 2018. Lecture Notes in Computer Science(), vol 11041. Springer, Cham. https://doi.org/10.1007/978-3-030-01201-4_34
Download citation
DOI: https://doi.org/10.1007/978-3-030-01201-4_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-01200-7
Online ISBN: 978-3-030-01201-4
eBook Packages: Computer ScienceComputer Science (R0)