Abstract
Segmenting filamentous structures in confocal microscopy images is important for analyzing and quantifying related biological processes. However, thin structures, especially in noisy imagery, are difficult to accurately segment. In this paper, we introduce a novel deep network architecture for filament segmentation in confocal microscopy images that improves upon the state-of-the-art U-net and SOAX methods. We also propose a strategy for data annotation, and create datasets for microtubule and actin filaments. Our experiments show that our proposed network outperforms state-of-the-art approaches and that our segmentation results are not only better in terms of accuracy, but also more suitable for biological analysis and understanding by reducing the number of falsely disconnected filaments in segmentation.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Filamentous structures are ubiquitous among biological systems and can be imaged by confocal fluorescence microscopy. Segmentation of these filamentous structures is important for understanding the mechanism of their formations and behavior.
There have been several attempts at segmenting filament structures based on traditional image processing techniques [1, 2, 6, 11, 13, 14]. Most of these traditional image processing approaches are based on photometric and geometric properties of filamentous structures. Because our aim is to study dynamic movements with high magnification microscopy, the images collected which contain higher levels of noise cause many traditional approaches to fail. Another common feature of these traditional methods is that appropriate parameter values need to be set accordingly to achieve a decent segmentation for different images [12, 14]. Hence, these methods work well only for a small data set, as it is cumbersome to adjust parameters for every individual image.
More recent approaches for general segmentation tasks are based on neural networks and have shown impressive performance for these types of tasks. Deep learning approaches have been applied to segment structures similar to filamentous structures [4, 5, 10]. Deep learning has been proven to work better than conventional image segmentation methods in the tasks mentioned above, but there is a limited number of works that segment filamentous structures in microscopy images. Though filamentous structures are similar to vesicular networks, retinal vessels, and cracks due to their piece-wise linear elements, the photometric and geometric properties of these structures vary significantly. Moreover, segmentation of filamentous structures in confocal microscopy images is complicated by optical blurring, noise, clutter, over exposure, and complex geometric properties such as overpass, convergence, and dense networks.
In this paper, we propose a new method utilizing a deep learning approach for automated segmentation of filamentous structures in microscopy images. Our work is built on U-net architecture [9], and we improve its performance for filament segmentation. Also, since there is no public data set for filament networks in microscopy images and it is time-consuming to annotate a large-scale data set for filamentous structures, we propose a semi-automatic annotation process based on a traditional segmentation method and a deep learning approach. By using this strategy, we create two data sets of microtubules and actin filaments.
The rest of this paper is organized as follows. Section 2 gives overview of work related to this paper. Section 3 details the process of data annotation, the architecture of network we proposed and training details. Section 4 describes our experiments, results and evaluations. Conclusions and future work will be given in Sect. 5.
2 Related Work
Filament Segmentation. There have been many works segmenting filamentous structures by using traditional methods like morphological approach [2], region-based approach [1, 6] and curve fitting approaches [11, 13, 14]. To make segmentation more robust to noise, Yue et al. [15] applied morphological operation and diffusion filtering algorithm to make the segmentation more robust to excessive white noise. Xu et al. [13] proposed a method called regulated sequential evolution. Combined with Stretching Open Active Contours (SOACs), they achieved more robust segmentation results. Based on SOACs method, Xu et al. [14] developed a convenient software tool called SOAX to segment filamentous structures. SOAX provides an easy-to-use user interface and is popular among researchers to do quantification analysis of biopolymer networks. However, SOACs method is a time-consuming method due to iterations. Moreover, to increase accuracy, it is necessary to adjust parameters depending on the type of filament and quality of the image [14]. As parameters for different images are mainly chosen empirically, it is hard for researcher to perform large-scale quantitative analysis. With appropriate parameters, many false predictions can be caused by other cell structures, over exposure, artifacts of images and so on. To improve the efficiency and accuracy, we want to apply deep learning approaches to filament segmentation. Though SOAX is not efficient in dealing with huge volumes of data and lacking accuracy in segmentation, it can assist our data annotation process and we will present the details in Sect. 3.
Vessel-like Structure Segmentation. Applying deep learning approaches to filamentous structure segmentation is rare, but there have been works using deep learning methods to segment vessel-like structures. Saponaro et al. [10] adapted U-net architecture to segment vesicular networks of fungal hyphae in macroscopic microscopy images. Fu et al. [5] utilized fully convolutional neural networks and fully-connected Conditional Random Field (CRFs) for retinal vessel segmentation in fundus image. Fan et al. [4] proposed a method for pavement cracks detection based on a convolutional neural network. Since the U-net [9] works well on vessel-like structure segmentation in microscopy images [10], we adapted and improved U-net architecture to segment filamentous structures on our data sets.
Neural Network Architecture. Ronneberger et al. [9] proposed U-net architecture which has achieved remarkable success in segmenting objects in biomedical microscopy images. This architecture is based on the idea of Fully-Convolutional Networks, and it contains contracting path to capture features and an expansion path to retrieve localization information. This architecture also applies overlap-tile strategy and allows the network training on relatively few training samples. Costa et al. [3] has applied this network to vessel segmentation in eye fundus images and it achieved a 0.9755 area under curve (AUC). This inspired us to apply this network on filamentous structures.
However, sometimes U-net creates small gaps where the hyphal networks are supposed to be continuous [10]. This can be caused by artifacts of the image or the U-net architecture itself. Newell et al. [8] introduce stacked hourglass networks for human pose estimation. This work has shown that repeatedly performing pooling and up-sampling with intermediate supervision can improve the performance of the network. To increase efficiency in a deep neural network, Huang et al. [7] introduces Dense Convolutional Network (DenseNet). In DenseNet, all layers with the same feature-map sizes are connected together in feed-forward fashion, which can encourage feature reuse, strengthen feature propagation and reduce the number of parameters [7]. Inspired by these works, our neural network architecture can take advantage from these networks to make our method more robust and avoid creating small gaps in filaments segmentation.
3 Method
Our goal is to create an efficient tool for filament segmentation in microscopy images. We utilize a semi-automatic scheme to annotate ground truth, and train our network on the data set.
3.1 Data Annotation
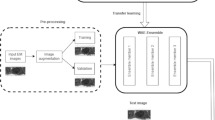
We propose a semi-automatic strategy to annotate filamentous structures in microscopy image to reduce the work of annotation. The main idea of this strategy is that we use SOAX [14] to segment images to obtain a weakly annotated mask. Then we use the single U-net module [9] (see Fig. 2), to train on these initially annotated training samples. We use this trained network to obtain segmentations for a larger amount of images. Based on the predicted segmentation masks, we ask domain experts to modify and correct these masks to finalize the ground truth. An example is shown in Fig. 1. We use the single U-net module instead of using our proposed network architecture (Sect. 3.2) to avoid the network overfitting on the weakly annotated masks.
There are two reasons why we don’t manually adjust the segmentation results of SOAX directly. First, the average time to run SOAX on a whole image at an approximate resolution of 2k by 2k pixels takes approximately 6 hours on high-end workstations. More false positive segmentations will be created due to noisy areas in the microscopy images, which will increase the work of manual modification. Therefore, we crop one image to several sub-images and then run them through SOAX, as shown in Fig. 1 (b) and (e). Since U-net takes patches as input, we only create training patches where there are SOAX segmentations and use 128 by 128 patches for training. After training, we use U-net to obtain the initial segmentation results of the entire image. From our experiments, the predicted initial segmentation results from single U-net module are more accurate than results of SOAX. Many false positive segmentations made by SOAX are removed, as shown in Fig. 1 (b), (c), (f), and (g). The IoU of results from SOAX and the single U-net module are 0.6189 and 0.7919 respectively when compared to the manually labeled images.
In total, we took 24 microscopy images with size of 122.03 \(\times \) 132.84 \(\upmu \text {m}\) (\(1400\times 1524\) pixels) and 17 slices in Z direction and obtained maximum intensity projection(MIP) on Z direction of these images. We cropped these 24 MIP images into 40 sub-images. By applying data augmentation strategy as in the work of Ronneberger et al. [9], we performed rigid transformation and \(\gamma \) correction on each valid patch and created 709800 training patches in total. We used these patches to train U-net and ran the trained network on 53 microscopy images including previous 24 images. In the end, domain experts manually checked and modified 53 full-resolution segmentation results, and each image took 10 to 25 min. We use 25 full-resolution images as the training set and 28 images as test set in all our experiments. We also create a data set for actin filaments with 10 microscopy images.
3.2 Network Description
In this paper, we build our network architecture based on U-net architecture [9], and we also adapted features of Stacked Hourglass Network [8]. Similar to U-net, we build up a module with contracting and expansion paths. Then we stack multiple modules end-to-end in a feed-forward fashion, which is similar to how Newell et al. [8] stack their hourglass network. The output of each module will be the input of next module. This allows the network repeatedly reevaluate previous prediction and features across all scales. The output of each U-net module will also go through a shared \(1\times 1\) convolutional layer to obtain a segmentation map. The loss function will take each intermediate output into consideration by assigning different weights to the loss values of the segmentation maps. This intermediate weighted supervision process can help each module optimize individually while attempting to improve upon the previous module’s segmentation. To help the network maintain the residual information that exists at intermediary stages, we add cross-connections between layers with the same feature-map sizes.
The network architecture is shown in Fig. 2. It contains three modules, each module includes two max pooling steps and two up-sampling steps. At each step, it contains two \(3\times 3\) convolutional layers followed by a rectified linear unit and one drop out layer with rate 0.2 inserted between the two convolutional layers to facilitate network generalization. For the contracting paths, each step will be followed by a \(2\times 2\) max pooling operation, and the number of feature channels will be doubled. For expansion paths, \(2\times 2\) up-sampling operation will be applied after each step halving the number of feature channels, and a concatenation operation on feature-map with matching size from all previous contracting paths. To obtain a segmentation map for each module, their output is connected to a shared \(1\times 1\) convolutional layer.
3.3 Training and Testing
For the annotation process, we used a single module of our proposed network, and we train this model for 20 epochs with a batch size of 64. On the microtubule data set we created, we train our proposed network for 15 epochs with a batch size of 64. The size of input patches is \(128\times 128\), and the number of training patches is 5032407. For each module, the numbers of feature channels are 32, 64, 128 for corresponding stages. All networks are trained using Adam optimizer with a learning rate of 0.0001 and a dice coefficient loss. Dropout rates of all dropout layers are set to 0.2. Due to GPU memory constrains, we implemented a generator to generate data batch-by-batch and fit our model. For our proposed network, there are multiple outputs. We compile the model and assign a weight of 0.2, 0.3 and 0.5 for the loss of first, second, and third module output separately. All experiments are conducted on a laboratory server with two NVIDIA GeForce Titan X (Pascal) GPUs.
4 Experiments
4.1 Evaluation
For evaluation, the Intersection over Union (IoU) method is applied, which is a commonly used metric. IoU metric can be very sensitive to pixel wise segmentation. Considering that our test data is manually annotated based on results of U-net and IoU metric can be biased, we propose an auxiliary metric called Skeletonized IoU (SKIoU) modified from IoU and defined as following:
This metric will ignore small misalignments and thickness of microtubules, as curvatures and length is much more important for domain experts. SKIoU will be much less sensitive and can be a fair metric for different methods.
We used both metrics along with opinions from domain experts to compare segmentation results of different approaches.
4.2 Segmentation Results on Microtubules
We have run 6 experiments on microtubules and results are shown in Table 1, and examples of segmentations results are shown in Fig. 3.
All neural networks perform better than SOAX software with regards to IoU and SKIoU. Our proposed network achieves highest score in IoU, and the network without cross connections achieves highest score in SKIoU. In general, the SKIoUs of all networks are very close to each other and our proposed network is slightly higher than single U-net Module with respect to IoU metric.
As shown in Fig. 3, networks with multiple modules stacked together outperforms single U-net module. Segmented results of our proposed network contain less disconnected microtubules and fragments than U-net, which is crucial for future quantification analysis. For example, These fragments will be considered as single microtubules in the future analysis and influence the final quantification result.
We also train single U-net for 30 epochs. The result is almost the same with results of 15 training epochs, which indicates that U-net can be hardly improved by training more epochs. Cross connections can improve the efficiency of our network. Table 1 showed that after 5 epochs, network with cross connections learns better than the one without cross connections.
4.3 Segmentation Results on Actin Filament
The structure of actin filaments is more complicated and denser than that of microtubules. Instead of training our network with actin filaments data, we applied the network trained with microbules data on actin filaments dataset. From Table 2, though our network achieves the highest IoU and SKIoU score, difference of SKIoU is rather small. However, it can be seen in Fig. 4 that U-net creates more fragments and gaps.
5 Conclusion and Perspectives
In this paper, we propose a new densely connected, stacked U-network architecture and also introduce a semi-automatic strategy to annotate filamentous structures. From our experiments, we show that the proposed deep network architecture not only achieves better accuracy but also produces segmentations that are more useful for biological analysis by reducing the number of falsely disconnected filaments and noise in segmentation than other state-of-the-art methods.
In the future, we will implement an application to quantify length, curvature and other information of filaments. Also, we will track the movement of filamentous structures over time, and by fusing the results of filaments and other structures like stromules, domain experts can better understand the formation and behavior of these structures.
References
Almi’ani, M.M., Barkana, B.D.: A modified region growing based algorithm to vessel segmentation in magnetic resonance angiography (2015)
Chang, S., Kulikowski, C.A., Dunn, S.M., Levy, S.: Biomedical image skeletonization: a novel method applied to fibrin network structures (2001)
Costa, P., et al.: Towards adversarial retinal image synthesis. arXiv preprint arXiv:1701.08974 (2017)
Fan, Z., Wu, Y., Lu, J., Li, W.: Automatic pavement crack detection based on structured prediction with the convolutional neural network. arXiv preprint arXiv:1802.02208 (2018)
Fu, H., Xu, Y., Wong, D.W.K., Liu, J.: Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 698–701. IEEE (2016)
Fuller, N., Aboudarham, J., Bentley, R.: Filament recognition and image cleaning on meudon h\(\alpha \) spectroheliograms (2005)
Huang, G., Liu, Z., Weinberger, K.Q., van der Maaten, L.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, p. 3 (2017)
Newell, A., Yang, K., Deng, J.: Stacked hourglass networks for human pose estimation. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9912, pp. 483–499. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46484-8_29
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Saponaro, P., et al.: DeepXScope: segmenting microscopy images with a deep neural network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 91–98 (2017)
Smith, M.B., Li, H., Shen, T., Huang, X., Yusuf, E., Vavylonis, D.: Segmentation and tracking of cytoskeletal filaments using open active contours (2010)
Xiao, X., Geyer, V.F., Bowne-Anderson, H., Howard, J., Sbalzarini, I.F.: Automatic optimal filament segmentation with sub-pixel accuracy using generalized linear models and B-spline level-sets. Med. Image Anal. 32, 157–172 (2016)
Xu, T., Vavylonis, D., Huang, X.: 3D actin network centerline extraction with multiple active contours (2014)
Xu, T., et al.: SOAX: a software for quantification of 3D biopolymer networks. Sci. Rep. 5, 9081 (2015)
Yue, G., Jiang, L., Liu, C., Yang, G., Ai, J., Chen, X.: Automated segmentation of microtubules in Cryo-EM images with excessive white noise. In: Kim, K.J., Joukov, N. (eds.) Information Science and Applications (ICISA) 2016. LNEE, vol. 376, pp. 339–348. Springer, Singapore (2016). https://doi.org/10.1007/978-981-10-0557-2_34
Acknowledgements
The National Institute of Health R01 grant GM097587 supported this work. Microscopy access was supported by grants from the NIH (P20 GM103446 and S10 OD016361).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, Y. et al. (2019). Densely Connected Stacked U-network for Filament Segmentation in Microscopy Images. In: Leal-Taixé, L., Roth, S. (eds) Computer Vision – ECCV 2018 Workshops. ECCV 2018. Lecture Notes in Computer Science(), vol 11134. Springer, Cham. https://doi.org/10.1007/978-3-030-11024-6_30
Download citation
DOI: https://doi.org/10.1007/978-3-030-11024-6_30
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11023-9
Online ISBN: 978-3-030-11024-6
eBook Packages: Computer ScienceComputer Science (R0)