Abstract
Accurate volumetric change estimation of glioblastoma is very important for post-surgical treatment follow-up. In this paper, an interactive segmentation method was developed and evaluated with the aim to guide volumetric estimation of glioblastoma. U-Net based fully convolutional network is used for initial segmentation of glioblastoma from post contrast MR images. The max flow algorithm is applied on the probability map of U-Net to update the initial segmentation and the result is displayed to the user for interactive refinement. Network update is performed based on the corrected contour by considering patient specific learning to deal with large context variations among different images. The proposed method is evaluated on a clinical MR image database of 15 glioblastoma patients with longitudinal scan data. The experimental results depict an improvement of segmentation performance due to patient specific fine-tuning. The proposed method is computationally fast and efficient as compared to state-of-the-art interactive segmentation tools. This tool could be useful for post-surgical treatment follow-up with minimal user intervention.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Glioblastoma is the most aggressive form of brain tumor and accounts for more than 80% of all primary brain malignancies [1]. Volumetric change estimation is very important for post-surgical treatment follow-up of patients with glioblastoma. Fully-automated segmentation of glioblastoma is difficult due to presence of necrosis and post-operative haemorrhages. There are different types of interactive tools existing in the literature based on the mode of outlining such as click-based [2], contour-based [3] and bounding box-based [4] and scribbling. Manual tumor segmentation is time consuming and the results are subject to inter- and intra-observer errors. In clinical practice, the diameter of glioblastoma in the biggest representative slice is often considered as the representative of the volume. However, the diameter is not a proper estimation of volumetric change as it is affected by the presence of post-operative cavities.
Several automatic and semi-automatic methods have been developed using support vector machines [5], random forests [6], markov random field [7] for segmentation of brain tumor. The majority of reported work on brain tumor segmentation use 2D filters for convolution [8, 9]. Structured predictions is implemented through convolution neural networks for better segmentation of brain tumor [9]. The U-Net [10] architecture considers the high-level features and the local appearance of tumor for better segmentation. Interactive segmentation methods have been implemented in several medical software [11, 12]. These methods require a relatively large amount of user interaction for segmentation of images with ambiguous boundaries.
In this paper, to reduce the time of user interaction for segmentation of glioblastoma, initial segmentation obtained from U-Net is refined by graph cut [13] algorithm and Smart paint [14]. The proposed method is novel as it includes an interactive step that levies a minimal effort on the expert who has to provide manual intervention at regions that have been mislabelled. It saves manual annotation of a few thousand pixels by the expert per patient. The work introduced in the paper are preliminary results along the direction of integrating energy minimization based post-processing in a semantic segmentation on a dataset with no available ground truth except for an expert’s judgement. Therefore, an attempt has been made to pick a fully convolution neural network based encoder-decoder architecture accepted by the medical community, such as U-Net and apply well understood post processing algorithm such as graph-cut to establish a baseline. The contributions of this paper are (i) design of a deep learning based interactive segmentation framework and (ii) implementation of patient-specific interactive refinement to deal with large content variations among images.
2 Materials and Methods
2.1 Database
A clinical MR image database (prospective research project, approved by local ethical committee) of brain tumors with world health organization (WHO) grade gliomas IV (glioblastoma) is used in this study. The database consists of longitudinal MR scans (post-operative) of 15 patients. The total number of MRI volumes of 15 subjects is 85. Contrast enhanced T1-weighted MR images were obtained from those 15 patients at defined time intervals. The boundaries of contrast enhanced tumour tissue is annotated by an expert radiologist using FsLeyes package [15] through slice by slice interaction. The average time spent for boundary annotation of each MRI volume is 12 min.
2.2 Interactive Segmentation Framework
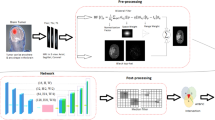
The block diagram of the proposed interactive segmentation framework is provided in Fig. 1. The availability of a sufficient number of annotated images is often a bottleneck in medical image analysis and U-Net [10] fills this gap by producing good results with a lower number of images. Therefore, the U-Net is used in the proposed framework. A set of cropped ROIs is used for training the U-Net and the trained model is named \(T_i\). Segmentation is performed for images of any MR instance of a patient based on \(T_i\) and the results are shown to user for interactive refinement by scribbling in foreground and background region. The pre trained model is updated based on the corrected mask created by interactive correction.
Architecture of U-Net. The proposed framework used U-Net [10] as it considers the high-level information and local appearance information of an object for better segmentation. It consists of an encoder and decoder paths. The architecture makes the use of strong data augmentation to use the available annotated samples more efficiently. The encoder path has five convolutional blocks and each block has two convolutional layers. The convolution is performed with a filter size of \(3\times 3\) and a stride of 1. Max pooling is performed with stride \(2\times 2\) to the end of every block except the last block. The size of the input images is \(64\times 64\) and it decreases from \(64\times 64\) to \(4\times 4\). In the decoder path, transpose convolution is applied in each layer with a filter size of \(3\times 3\) and stride of \(2\times 2\). In each block, the size of the feature maps is doubled but the width of the feature maps is decreased by factor of 2. Therefore, the size of feature maps increases from \(4\times 4\) to \(64\times 64\). High-level information is represented at up-sampling blocks, and low-level features are transferred through skip connections.
Off-Line Training of U-Net. Patches of size \(64\times 64\) are extracted from axial slices of brain MR images for the training of U-Net. Rotation and flip are used to increases the number of training images. In training of U-Net, a stochastic gradient-based optimization [16] (Adam) is applied to minimize the cross-entropy based cost function. The cross-Entropy loss function has been used for this purpose due to its efficiency in binary classification. Adam utilizes the first and second moments of gradients for updating and correcting moving average of the current gradients. The learning rate for the Adam optimizer is set to 0.0001, weights of background and foreground are initialized as 1 : 10, and training were performed up to 60, 000 iterations. Dropout [17] is used to reduce over-fitting. A training and validation dataset pair \((I_r,I_{r_g})\); \((I_v,I_{v_g})\), consisting of MRI images from patients with post-operative glioblastoma is used to train a CNN based on vanilla U-Net architecture from scratch, to obtain a preliminary CNN model \(T_i\) (stage-1 of Fig. 1).
Update of Initial Segmentation Obtained by U-Net. The output of U-Net is updated using a graph cut algorithm [13]. In case of interpreting an images as a graph, its pixels are considered as vertices and image edges are considered as graph edges. The optimal solution results in a cut on the graph that minimizes the energy, which corresponds to maximizing the flow along those edges. The underlying idea is to formulate the refined segmentation of image I as an energy minimization problem as given in the Eq. (1) resulting in a binary image B

where \(L_{ij}^t,L_{ij}^b,C_{ij}^h, C_{ij}^v\) are costs associated with labeling a pixel indexed (i, j) as belonging to tumor, brain, smoothness along horizontal and vertical directions, respectively. Labeling costs are governed by Eq. (2)
where the labeling cost \(L_{ij}^t, L_{ij}^b\) are interpreted as a probabilities \(P(t_{ij} | I_{ij}; W)\), \(P(b_{ij} | I_{ij}; W)\) of the pixel \(I_{ij}\) at (i, j) being labeled as tumor or brain respectively. The probabilities are obtained by applying a Softmax function over the class scores \(l_{t_{ij}}, l_{b_{ij}}\) derived from the U-Net model with weights W for tumor and brain classes respectively.
Smoothness cost is governed by Eq. (3)

which encourages label consistency on either sides of the edge \(E_{ij}\) for an associated cost \(\phi \). Details on graph-cut based semantic segmentation improvement can be found in [18].
Patient-Specific Refinement of Pre-trained Model. The procedure of interactive refinement is described in stage-2 and stage 3 of Fig. 1. A test dataset \(I_{s_1}\) for which no ground truth exists, is segmented using \(T_i\) to obtain segmented output \(I_o\). A graph-cut is applied to the class probability maps from \(T_i\) to get an output \(I_g\).
-
Though \(I_g\) can be directly used for interactive segmentation, it is empirical found that using \(I_g \odot I_o\) (Hadamard product of the two images) helps reduce false positives.
-
\(I_g \odot I_o\) is used in an interactive tool by an expert to obtain the ground truth segmentation for \(I_{s_1}\) as \(I_{sg_1}\).
The pre-trained U-Net model \(T_i\) from Stage-1 is re-trained on \((I_{s_1}, I_{sg_1})\) to get a refined model \(T_{i+1}\). Stages 2, 3 can be iterated to get better models using \(T_{i+1}\). Images of a particular instance of longitudinal data is used for patient specific fine-tuning and another instance of that patient \((I_t,I_{t_g})\) is used for testing to report the dice similarity coefficient (DSC).
3 Experiments, Results and Discussion
Training of U-Net has been performed in Linux environment using a 11 GB GPU (Zotac GeForce GTX 1080 Ti) on a system with Core-i7 processor and 32 GB RAM. The network architecture is implemented in Python using the PyTorch library. DSC is considered as performance metric for evaluation of segmentation results, as defined below:
where, \(N_{tp}\), \(N_{tn}\), \(N_{fp}\) and \(N_{fn}\) is number of true positive, true negative, false positive and false negative pixels, respectively.
The segmentation results is evaluated by considering 15-fold cross-validation. A trained medical student, who is familiar with the glioblastomas database, has corrected the initial segmentation using Smart paint [14] for network update. Segmentation was also performed using several interactive segmentation tools such as 3D paint and brush tool of ITK-Snap [19] and 3D-Slicer [20] for comparative study. Box plots of DSC achieved by several methods are shown in Fig. 2. The result shows substantial improvement over existing methods. The proposed approach is straight forward and simple. It is an intentional choice to explore the improvement of segmentation based on subject specific refinement. The max-flow algorithm plays an important role in improving segmentation accuracy in the boundary region. The improvement of mean DSC is around 3% due to the inclusion of max-flow algorithm. Moreover, there is reduction of computational time if max-flow algorithm as it is applied on the result of U-Net. Interactive refinement is required in more than 90% of the cases. Therefore, max-flow can not be avoided. Comparative study of interactive segmentation time required by the expert is presented in Table 1. Time of interactive segmentation to achieve a sufficiently high accuracy is noted for all methods. The proposed method takes less time to achieve acceptable segmentation result.
The initial segmentation results of U-Net and result after patient-specific interactive refinement is shown in Fig. 3. Initial segmentation by U-Net is poor due to the presence of a necrosis and post-operative haemorrhage. It is observed that incorporating certain image features related to the patient, improves the segmentation results.
4 Conclusion
A patient-specific interactive refinement method is implemented using U-Net based fully convolutional network. The proposed interactive segmentation framework can provide both efficient and robust segmentation compared to other interactive tools. The network is able to learn patient-specific features from the feedback provided by an expert user and results better segmentation for rest of the longitudinal MR images of the same subject. This method will be helpful in treatment follow-up analysis in neuroradiology.
References
Ostrom, Q.T., et al.: CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro-oncology 17(Suppl. 4) (2015)
Haider, S.A., et al.: Single-click, semi-automatic lung nodule contouring using hierarchical conditional random fields. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 1139–1142. IEEE (2015)
Xu, C., Prince, J.L.: Snakes, shapes, and gradient vector flow. IEEE Trans. Image Process. 7(3), 359–369 (1998)
Rother, C., Kolmogorov, V., Blake, A.: GrabCut: interactive foreground extraction using iterated graph cuts. In: ACM Transactions on Graphics (TOG), vol. 23, pp. 309–314. ACM (2004)
Lee, C.-H., Wang, S., Murtha, A., Brown, M.R.G., Greiner, R.: Segmenting brain tumors using pseudo–conditional random fields. In: Metaxas, D., Axel, L., Fichtinger, G., Székely, G. (eds.) MICCAI 2008. LNCS, vol. 5241, pp. 359–366. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-85988-8_43
Tustison, N.J., et al.: Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR. Neuroinformatics 13(2), 209–225 (2015)
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015)
Lyksborg, M., Puonti, O., Agn, M., Larsen, R.: An ensemble of 2D convolutional neural networks for tumor segmentation. In: Paulsen, R.R., Pedersen, K.S. (eds.) SCIA 2015. LNCS, vol. 9127, pp. 201–211. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-19665-7_17
Dvorak, P., Menze, B.: Structured prediction with convolutional neural networks for multimodal brain tumor segmentation. In: Proceedings of the Multimodal Brain Tumor Image Segmentation Challenge, pp. 13–24 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Armstrong, C.J., Price, B.L., Barrett, W.A.: Interactive segmentation of image volumes with live surface. Comput. Graph. 31(2), 212–229 (2007)
Cates, J.E., Lefohn, A.E., Whitaker, R.T.: GIST an interactive, GPU-based level set segmentation tool for 3D medical images. Med. Image Anal. 8(3), 217–231 (2004)
Boykov, Y., Kolmogorov, V.: An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans. Pattern Anal. Mach. Intell. 26(9), 1124–1137 (2004)
Malmberg, F., Strand, R., Kullberg, J., Nordenskjöld, R., Bengtsson, E.: Smart paint a new interactive segmentation method applied to MR prostate segmentation. In: MICCAI Grand Challenge: Prostate MR Image Segmentation 2012 (2012)
Paul, M.: FSLeyes. https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLeyes
Kingma, D., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Hinton, G.E., Srivastava, N., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.R.: Improving neural networks by preventing co-adaptation of feature detectors. preprint arXiv:1207.0580 (2012)
Ayyalasomayajula, K.R., Brun, A.: Historical document binarization combining semantic labeling and graph cuts. In: Sharma, P., Bianchi, F.M. (eds.) SCIA 2017. LNCS, vol. 10269, pp. 386–396. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59126-1_32
Yushkevich, P.A., Gao, Y., Gerig, G.: ITK-SNAP: an interactive tool for semi-automatic segmentation of multi-modality biomedical images. In: 2016 IEEE 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), pp. 3342–3345. IEEE (2016)
Fedorov, A., et al.: 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30(9), 1323–1341 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Dhara, A.K. et al. (2019). Segmentation of Post-operative Glioblastoma in MRI by U-Net with Patient-Specific Interactive Refinement. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11383. Springer, Cham. https://doi.org/10.1007/978-3-030-11723-8_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-11723-8_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11722-1
Online ISBN: 978-3-030-11723-8
eBook Packages: Computer ScienceComputer Science (R0)






