Abstract
This paper presents a novel Multipath Densely Connected Convolutional Neural Network (MDCNN) for automatically segmenting glioma with unknown sizes, shapes and positions. Our network architecture is based on the Multipath Convolutional Neural Network [21], which considers both local and contextual patches of segmentation information, including original MRI images, symmetry information and spatial information. Motivated to reduce the feature loss induced by under-utility of feature maps, we propose to fuse feature maps from original local and contextual paths at three different units and introduce three more densely connected paths. Consequently, three auxiliary segmentation paths together with original local and contextural paths forms the complete segmentation network. The model’s training and validation are performed on the BraTS2017 dataset. Experimental results demonstrate that the proposed network is capable to effectively extract more accurate tumor locations and contours with improved stability.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Glioma is a type of tumor that occurs in the brain and spinal cord, and it accounts for almost 80% of primary malignant brain tumors [20]. Statistics show that brain and other central nervous system (CNS) tumors are the second most common cancers in children and adolescents [6]. What’s worse is that brain tumor in children easily leads to related diseases, such as visual impairment and hemiplegia.
Last few decades have witnessed significant improvement in clinical neuro-oncology treatment, including neurosurgery surgery, chemotherapy, radiotherapy, physiotherapy and etc. [23]. Medical images, such as computed tomography (CT), ultrasound and magnetic resonance imaging (MRI), act as an irreplaceable role in these techniques, since they can provide indispensable information for diagnosis and treatment. Comparing with other medical imaging modalities, MRI can form clear and complete pictures of the lesions from various image sequences [3]. This advantage makes MRI being widely used in hospitals and clinics for medical diagnosis and treatment of many diseases [7].
Whereas MRI brings great conveniences for tumor diagnosis, it’s not straightforward to accurately segment MRI images for extracting the patient-specific important clinical information, and their diagnostic features [15, 24]. Moreover, due to the large amount of data, it is time consuming and tedious for the radiologists to manually annotate and segment the data, resulting in limited usage for objective quantitative analysis [3]. To tackle this issue, great efforts are put for automatic brain tumor segmentation. Considerable research has been conducted for glioma segmentation in MRI images, it’s still a challenge task to achieve satisfied results due to several inherent difficulties:
-
It requires large dataset to learn stable characteristics feature map because of the uncertainty in shape and characteristics of glioma.
-
It’s difficult to balance between large context features and local detail features because of various region sizes of different tissue distributions.
-
It easily causes class imbalance due to inhomogeneity of label classification.
Abundant of solutions are proposed aiming at solving this problem from different perspectives. To reduce continuous resolution reduction, Long et al. [14] propose to segment images using Fully Convolutional Networks (FCN) and achieve pixel level segmentation on images with various scales. However, the results has limited accuracy because max-pooling and sub-sampling would reduce the resolution of feature maps. To solve this problem, SegNet [1] and U-Net [19] add up-sampling layers (decoder network) after down-sampling layers (encoder network) to map low resolution feature maps to full input resolution feature maps. This architecture improves segmentation accuracy due to more features retained from training data. U-Net has been successfully adopted for real brain tumor segmentation [4].
With the depth of the network deepens, SegNet and U-Net would “get lazy” in feature extraction, i.e., it would easily lose abstract features. ResNet [9] are proposed to improve the learning capability of deeper features with deeper models. It reformulates the layers as learning residual functions with reference to the layer inputs, rather than learning unreferenced functions.
By introducing direct connections between any two layers with the same feature-map size, DenseNet [11] can naturally scale to hundreds of layers without exhibiting optimization difficulties. Moreover, DenseNet requires substantially fewer parameters and less computation because of the adoption of hyperparameter settings optimized for residual networks.
Meanwhile, dual path cascade architecture also has achieved good results on brain tumor segmentation task [8]. [12] simultaneously exploits local features and more global contextual features than traditionally CNN architecture and obtains competitive performance with much higher efficiency. [21] further improves the network of [12] by considering spatial information and high asymmetry property caused by the contrast between healthy brain tissue and tumor tissue.
Some other researchers design segmentation networks by integrating advantages from multiple different networks. [18] proposes a natural image segmentation algorithm by introducing a special residual function with a path of multiple stream. [11] employed this method on brain tumor segmentation and achieved appealing results. [16] and [17] integrate several components into CNNs, including residual structure, up-sampling, down-sampling and symmetric/asymmetric convolution structure. Therein, E-Net [16], significantly improve efficiency with competitive results while [17] performs better in boundary segmentation by enlarging perception of different convolution sum. In addition to residual structure, [5] continuously fuses feature information from different convolutional layers to retain detail features and reduce in-balance problem.
In this paper, we propose a Multi-path Densely Connected Convolutional Neural Network (MDCNN) which fuses information in multiple stages to improve the stability and reduce feature loss, and define a loss function to support multi-path segmentation.
2 Database
Data from BarTS Challenge 2017 [2], which is composed of training, validation and test datasets, is employed for our experiments. Since the date of competition has expired and unable to get the test datasets, we only use the training and validation dataset. The training dataset contains MRI data of 285 subjects, 210 with high grade glioma (HGG) and 75 with low grade glioma (LGG). Each training subject provides four MRI modalities, namely T-weighted(T1), post-contract T1-weighted(T1ce), T2-weighted(T2) and T2 Fluid Attenuated Inversion Recovery(FLAIR). In addition, four ground truth classes are given by images with resolution 240 \(\times \) 240 \(\times \) 155. The four classes represent necrotic and non-enhancing tumor (NCR and NET, label 1), peritumoral edema (ED, label 2), enhancing tumor (ET, label4) and everything else (label 0), respectively.
80% of the training dataset (168 HGG and 60 LGG subjects) is used for training and the remaining 20% (42 HGG and 15 LGG subjects) is used for validation. At last, the validation dataset without GT from 46 patients is used to test the proposed network.
3 Methods
Our method is motivated to make full usage of features for better segmentation performance by integrating advantages from multi-path Convolutional Neural Network [21] and densely connected multi-scale feature maps [11]. The work of [21] uses multi-path to encode both contextual and local information and proposes specific pre-processing and post-processing procedures for better performance. However, it hasn’t taken full advantage from feature maps. Multi-scale feature maps and dense connectivity are introduced to further improve the segmentation accuracy. This shares similar idea as MSDNET [10], which is originally proposed to classify images with strong randomness and large variation range. So, the proposed method not only fuse the feature maps at the final stage of the two paths, but also introduce fusion in multiple stages and add dense connection between the fused feature maps. Even local and contextural paths may lose some features, the segmentation network of fusion feature can compensate with different features and thus can improve the performance. This architecture can continuously strengthen the transmission between feature maps, thus making more efficient usage of feature maps and reducing vanishing-gradient [10, 11]. Even with more parameters, the network is more stable and has stronger learning ability.
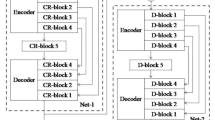
Figure 1 illustrates the network structure. The network extracts both large contextual patches and local patches as multi-path CNN does, so it can well retain contextual information. On the basis work of [21], symmetry information is used to differentiate healthy tissue and tumor tissue. To facilitate re-usage of feature maps for fusion in the same scale, we change the patch sizes on the contextual region path and on local region path in multi-path CNN to 48 \(\times \) 48 \(\times \) 11 and 20 \(\times \) 20 \(\times \) 4, respectively. In addition to convolution operation in each path, we fuse feature maps from large region path and local region path. The fusion operation can make better usage of feature maps. The first fusion of feature maps from two paths is conducted when the size of feature map is 16 \(\times \) 16 (Illustrated in the red part in Fig. 1). The fusion generates the first auxiliary densely connected segmentation path. In the same manner, the second and third auxiliary densely connected segmentation path are generated by fusing the feature maps with size 12 \(\times \) 12 and 8 \(\times \) 8 (Illustrated in the blue and yellow regions respectively). Further, first auxiliary layer with size 16 \(\times \) 16 is proceeded to layer with size 8 \(\times \) 8 (See the yellow rectangle in Fig. 1) and fused to the second auxiliary densely connected segmentation path. Similarly, the second auxiliary layer is proceeded to layer with size 4 \(\times \) 4 and fused to the third auxiliary layer. Each fused feature map will generate an auxiliary densely connected segmentation path with normal convolution and FCN. We also tried to introduce fusion on the layer with patch size 4 \(\times \) 4 and found that it can’t significantly improve the performance but introduces more parameters. So we finally chose to use three fusion segmentation paths for the best performance. Except for the parameters shown in Fig. 1, the layers between 48 \(\times \) 48 \(\times \) 11 to 16 \(\times \) 16 \(\times \) 64 and the FCN parameters on the main path and branches are given in Table 1.
After several convolution operation and dropout regularization [22], contextural and local feature vectors are concatenated and put forward for another FC layer. Finally, softmax function is adopted for classification. In the training process, we employ DisturbLabel regularization, which randomly set GT to 0 or 1, to improve the stability.
Besides, considering that brain tumor segmentation has relatively lower randomness, we introduce the idea of dense connection [10] to our net. In addition, the segmentation results from four paths, i.e., main path and three densely connected paths, are weighted by 0.25 summed up for the final loss. The loss function is:
in which, S is the number of branches, X is the input with size \([N,h*w*d]\), \(\omega _s\) is the weight, \(\lambda _{1}\) and \(\lambda _{2}\) are the weights correlating to the ratio of the two cases. \(L_s\) is the loss function defined for each branch:
in which, N is the number of samples in the batches, \(q_{i,c}\) means the segmentation results with respect to label c, and \(Z_{i,c}^{gt}\) is the corresponding label. In this paper, we simultaneously consider two classification cases, namely two classes (with label 0 or 1) and four classes (with label 0,1,2,4), to further enhances the learning capability.
4 Results
The method is implemented with Python and trained with TensorFlow. The hardware platform is PC with NVIDIA Titan X (12 GB) GPU and Intel\(^{\textregistered }\) Xeon(R) CPU E3-1231 v3 @ 3.40 GHz x8 32 GB). The detailed parameters in network are given in Fig. 1 and Table 1.
In pre-processing, all data are clipped into range \([-2.0, 2.0]\). The sizes of contextual patches and local patches are 48 \(\times \) 48 and 20 \(\times \) 20, respectively. Dropout probability used during the entire training process is 0.5. Weights for the two classification cases (\(\lambda _{1}\) and \(\lambda _{2}\)) are 0.2 and 0.4. These parameters are empirically chosen based on [21]. In the training process, Adama optimization algorithm [13] is used to update weights. Different parameters are used in Adama optimizers in different phases.
Two phases are defined in training, rather than 3 phases use by [21]. The two phases correlate to pre-training and fine training. During the first phase (iteration \(\le \) 9000), we randomly chose data of 5 subjects and extract 50 samples for each class in each iteration. During the second phase (9000 < iteration \(\le \) 13000), 10 subjects and 25 samples for each class are randomly chosen in each iteration. \(10^{-4}\) and \(10^{-5}\) are chosen as the learning rate for these two phases.
In post-processing, thresholding is first applied to convert the output, which represents the probability of being different regions, to integer values matching with the GT data type. The thresholding values for being tumorous or being enhancing tumor are 0.905 and 0.45, respectively. In addition, after thresholding, connected components with less than 3000 voxels will be re-classified as healthy.
Our experiment is conducted on various label combination, including whole tumor region with labels 1, 2, 4 (WT), core tumor region with labels 1 and 4 (TC), and enhancing tumor region with label 4 (ET). We adopt 20% of the training dataset (Data of 57 subjects) and validation dataset (Data of 46 subjects) in validation and testing, respectively. Besides, Dice score and 95% Hausdorff are used for measurement. The results all we show in the tables are received after uploading our segmentation results to the official website. Since BraTS2017 testing dataset is not available, we use the validation dataset for testing. The evaluation results of validation dataset are also received after uploading our segmentation results to the official website, because we have no the Ground Truth.
Table 2 shows the results of our method and that of [21] both with post-processing. The thresholding of being tumorous or being enhancing tumor used by [21] are 0.916 and 0.4. We can find that our method achieve better performance for all segmentation tasks (WT, TC and ET) than [21]. On the training dataset, the presented method gains nearly 10% improvement, and on the validation dataset, we achieve 3% improvement in average.
Table 3 compares the results without post-processing from our method and that from [21]. The comparison is conducted on the testing dataset from training data. It shows our method achieves 6.7% improvements in average.
Tables 4 and 5 are specified segmentation results on LGG and HGG training dataset with or without post-processing. It can be observed that our results with or without post-processing are prior to the result from [21]. Besides, ET segmentation results on LGG dataset are significantly poorer than that on HGG dataset. The main reason might be that LGG dataset is much smaller than HGG dataset. Thus, collecting more LGG training subjects would improve the performance.
Several segmentation results are illustrated in Figs. 2 and 3. It can be noticed that, post-processing can clearly improve the segmentation results on those subjects with more smooth boundaries. In this case, post-processing can effectively filter out redundant false details, thus improving the performance. Meanwhile, segmentation for those subjects with bumping region boundaries rely more on the segmentation capability of the network, and the promotion from post-processing would be small.
5 Conclusions and Future Work
This paper presents a novel Multi-path Densely Connected Convolutional Neural Network for glioma segmentation. By introducing densely connections on fused feature maps, the proposed model can well retain both abstract features and specific features for deep network. Thus, the network becomes stable and with higher learning capability. Experiments indicate that our method achieves the highest performance for various segmentation tasks. In future, we intend to reduce the memory cost of the network and extend the framework to more clinical applications.
References
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Bakas, S., et al.: Advancing The Cancer Genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017)
Bauer, S., Wiest, R., Nolte, L.-P., Reyes, M.: A survey of MRI-based medical image analysis for brain tumor studies. Phys. Med. Biol. 58(13), R97 (2013)
Dong, H., Yang, G., Liu, F., Mo, Y., Guo, Y.: Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. In: Valdés Hernández, M., González-Castro, V. (eds.) MIUA 2017. CCIS, vol. 723, pp. 506–517. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-60964-5_44
Fidon, L., et al.: Generalised Wasserstein dice score for imbalanced multi-class segmentation using holistic convolutional networks. arXiv preprint arXiv:1707.00478 (2017)
Gittleman, H.R., et al.: Trends in central nervous system tumor incidence relative to other common cancers in adults, adolescents, and children in the United States, 2000 to 2010. Cancer 121(1), 102–112 (2015)
Gordillo, N., Montseny, E., Sobrevilla, P.: State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 31(8), 1426–1438 (2013)
Havaei, M., et al.: Brain tumor segmentation with deep neural networks. Med. Image Anal. 35, 18–31 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Huang, G., Chen, D., Li, T., Wu, F., van der Maaten, L., Weinberger, K.Q.: Multi-scale dense networks for resource efficient image classification. arXiv preprint arXiv:1703.09844 (2017)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017)
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015)
Paszke, A., Chaurasia, A., Kim, S., Culurciello, E.: ENet: a deep neural network architecture for real-time semantic segmentation. arXiv preprint arXiv:1606.02147 (2016)
Peng, C., Zhang, X., Yu, G., Luo, G., Sun, J.: Large kernel matters-improve semantic segmentation by global convolutional network. arXiv preprint arXiv:1703.02719 (2017)
Pohlen, T., Hermans, A., Mathias, M., Leibe, B.: Full-resolution residual networks for semantic segmentation in street scenes. arXiv preprint (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Schwartzbaum, J.A., Fisher, J.L., Aldape, K.D., Wrensch, M.: Epidemiology and molecular pathology of glioma. Nat. Rev. Neurol. 2(9), 494 (2006)
Sedlar, S.: Brain tumor segmentation using a multi-path CNN based method. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 403–422. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_35
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
Tabatabai, G., et al.: Molecular diagnostics of gliomas: the clinical perspective. Acta Neuropathol. 120(5), 585–592 (2010)
Yamahara, T., et al.: Morphological and flow cytometric analysis of cell infiltration in glioblastoma: a comparison of autopsy brain and neuroimaging. Brain Tumor Pathol. 27(2), 81–87 (2010)
Acknowledgment
This work was supported in part by the Shenzhen Science and Technology Program (No. JCYJ20160429190300857 and No. JCYJ20170413162617606), Research Grants Council of the Hong Kong Special Administrative Region (Project No. GRF 14203115) and the National Natural Science Foundation of China (Grant No. 61802385). Yinling Qian and Weixin Si are the corresponding authors.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, C. et al. (2019). Multipath Densely Connected Convolutional Neural Network for Brain Tumor Segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11383. Springer, Cham. https://doi.org/10.1007/978-3-030-11723-8_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-11723-8_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11722-1
Online ISBN: 978-3-030-11723-8
eBook Packages: Computer ScienceComputer Science (R0)