Abstract
Electrocardiogram (ECG) is an essential source of information for heart diseases classification. Hence, it is used by the cardiologist to diagnose heart attacks and detect the abnormalities of the heart. The automatic classification of the ECG signals is playing a vital role in the clinical diagnosis of heart diseases. In this paper, an end-to-end classification method is proposed using 1D Convolution Neural Networks (CNN) to extract the important features from the input signals and classify it automatically. The main advantage of CNN compared to the related work methods is that it gets rid of the hand-crafted features by combining the feature extraction and the classification into a single learning method without any human supervision. The proposed solution consists of data filtering, dynamic heartbeat segmentation, and 1D-CNN consisting of 10 layers without the input and the output layers.
Our experimental results on 14 classes of the public MIT-BIH arrhythmia dataset achieved a promising classification accuracy of 97.8% which outperforms several ECG classification methods.
Similar content being viewed by others
Keywords
1 Introduction
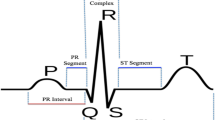
ECG is used to measure the electrical activity of the heart over time by putting a set of electrodes on the body surface such as chest, arms and neck. These electrodes can detect the electrical changes in the heart. ECG signals are mainly consisting of three complex waves such as P, QRS, and T waves. The P wave shows the atria contractions, while QRS complex shows the ventricular contractions and the T wave reflects the electrical activity produced while recharging the ventricles for the next contractions [1]. Hence, different cardiac activities can be represented by these complex waves and their study plays a vital role in the diagnosis of different arrythmias [2]. However, it is not an easy task for a cardiologist to analyze huge amount of ECG data due to its complexity as well as time requirements [3]. Moreover, the life threating types of arrhythmias need early and accurate detection [4].
There are two main categories for arrhythmias; the first one causes cardiac arrest and sudden death, such as ventricular fibrillation and tachycardia [5, 6]. The second category, which is our focus in this paper, needs care to avoid deterioration but it is not as dangerous as life-threatening diseases in the first category [7]. Detection and classification of arrythmias require the classification of heartbeats. We can determine the heart rhythm category by recognizing the classes of consecutive heartbeats [8]. Beat by beat human-based classification is a time-consuming task and too difficult of a process. Therefore, the automation process of ECG analysis is very important to discover cardiac disorders which need immediate medical aid in clinical situations, and it also will save a lot of time and efforts for the cardiologist.
In the recent past, different approaches have been presented for automatic classification of heartbeats such as Support Vector Machines (SVM) [9, 10], Back Propagation Neural Networks (BPNN) [11], regression neural networks [12] and Recurrent Neural Networks (RNN) [31].
Deep learning has advanced rapidly since the start of the current decade. Recently, its techniques have shown promising results and demonstrated state-of-the-art performances in tremendous tasks, due to the availability of a huge amount of data and the dramatic increase in the current computational power specially in the modern GPUs, in various fields such as Bioinformatics [13] and Medical diagnosis [14]. One of the advantages of the deep learning techniques is its structure in which both feature extraction and classification stages are performed together without requiring hand-crafted features, which is called end-to-end learning [15].
In this paper, the proposed solution consists of two steps; preprocessing and classification steps. Firstly, the preprocessing step consists of data filtering and dynamic heartbeat segmentation. Data filtering is utilized to remove the noise from the ECG records as well as improving the quality of the signals and increasing the Signal-to-Noise Ratio (SNR). Heartbeats are dynamically segmented to avoid heart rate variability. Secondly, a robust end-to-end method is used to extract the most important features and classify them into 14 classes of the public MIT-BIH arrythmia dataset, using Convolution Neural Networks (CNN), with superior performance than the existing studies.
The remainder of this paper is organized as follows: in section two, the related work is provided. Section three discusses the proposed architecture and methodology in detail. Section four represents the achieved results and finally, the conclusion and the future work are provided in section five.
2 Related Work
The conventional way of ECG heartbeat classification is to develop an algorithm to extract the most important features from the signal and then choose an appropriate method to be used in the classification stage. Undoubtedly, there are many researchers in the literature who have done a lot of studies of such way for ECG classification.
Yu [16] used Independent Component Analysis (ICA) to extract the features and the classification is done between eight classes using Neural Networks and they have achieved accuracy of 98.71%, while in [17] the classification is done between four classes using Support Vector Machine (SVM) and the features are extracted using Discrete Wavelet Transform (DWT) and they have achieved accuracy of 98.39%. In [18] the authors combined the ICA with DWT features with the use of Probabilistic Neural Networks (PNN) for classification between five classes and an accuracy of 99.28% is achieved.
Yazdanian et al. [20] have considered five different classes and they have achieved accuracy of 96.67% using a set of wavelet, morphological and time-domain features. Furthermore, The same five classes have been considered by Martis et al. [19] and an accuracy of 98.11% is achieved with comparison for different approaches for feature extraction such as Principal Component Analysis (PCA) and DWT.
In [21] the feature set is a combination of linear and non-linear features to improve the classification of ECG data, SVM is used for classification between five classes and they have achieved 98.91% recognition accuracy. The classification is done in [22] between 13 classes and the features are generated using two methods; the first method is the Higher Order Statistics (HOS) and the second is the Hermite characterization, the achieved classification accuracy is 98.18%. Khazaee [23] used morphological and time-duration features to represent each beat and the classification is done using SVM between three different classes (Normal, Premature Ventricular Contraction and Others) and an accuracy of 99.9% is achieved. In [24], non-linear transformation is utilized followed by PCA to reduce the dimensionality of the features. Thereafter, the reduced feature vector is fed into SVM for classification, five beats are considered and an accuracy of 98.70% is achieved.
Finally, in [25] they proposed a two-stage hierarchical method to classify the heartbeats of MIT-BIH arrythmia dataset into 15 classes. DWT is used to extract the morphological features and then reduced by PCA. After that, these features are concatenated with four RR features and fed into SVM classifier, an overall accuracy of 94.94% is achieved across ten trials.
To summarize, the most widely used arrythmia dataset in the literature is MIT-BIH. There are many researchers who use fixed window to segment the heartbeats. PCA, ICA and DWT are the most common methods used to extract the features and SVM is the dominant method in the classification stage. Finally, most of the existing studies consider few classes. The aim of this paper is to get rid of the hand-crafted methods of feature extraction used in the literature by using one of the most promising techniques in the deep learning (CNN). In addition, more arrythmia classes are considered.
3 Methodology
This section discusses the used approaches for preprocessing and classification. Figure 1 describes the proposed method which has two main steps; data preprocessing and classification. A detailed description of the proposed method will be discussed in the following sub-sections.
3.1 Preprocessing
This step is essential to improve the signal-to-noise ratio as well as the classification accuracy. The noise is reduced by removing both low and high frequencies out of the ECG signal. Butterworth bandpass filter is applied with a range 0.5–40 because this range contains the most valuable information of the signal [28]. The effect of such filter is shown in Fig. 2. Using the R peak locations which are associated with the database in an annotation file, each ECG record is segmented into multiple heartbeats, and each heartbeat contains P wave, QRS complex, and T wave. Due to the difficulty of detecting the start and the end for each beat, a fixed segmentation method is usually applied. However, it is not always reliable because that assumption cannot consider the variations of the heart rate. Hence, a dynamic segmentation strategy is utilized to overcome the variability of the heart rate as proposed in [10]. The dynamic segmentation strategy counts the number of samples before and after each R peak according to the duration between the current and previous R peaks (RR previous) in addition to the duration between the current and the next R peaks (RR next). After that, it takes a portion from each interval that ensures to include all three complex waves. Such method is invariant to the heart rate variability. Finally, all heartbeats are resampled to have 300 samples per each heartbeat as done in [3, 25].
3.2 Convolution Neural Networks
Convolution Neural Networks (CNN) is mainly composed of feature extraction and classification stages. The feature extraction stage is responsible for extracting the most useful information from the ECG signals automatically. These features are fed into the classification part to accurately classify these features to one of the target classes. The feature extraction part consists of convolution and pooling (down sampling) layers. Convolutional layers are responsible to extract features from the input data by applying the convolution operation between the input signal and the convolution kernels. Finally, the result is calculated by passing the computed value to an activation function to add non-linearity to these features. Convolutional layer’s output can be represented by Eq. 1:
Where \( y_{k}^{l} \) is the output vector of the \( l^{th} \) layer with \( k^{th} \) convolution kernel and \( x_{k}^{l} \) is the input vector, while \( w_{k}^{l} \) is the weights of the convolution kernel and \( b_{k}^{l} \) is the bias coefficient and \( \phi \) is the activation function.
Max pooling is applied after the convolutional layer to reduce the dimensionality and preserve the useful information. In the output layer, the Softmax function is used to give a probability to each class of the target classes. The loss is computed by comparing the output with the target vector according to the cross-entropy function in Eq. 2:
Where \( y \) is the output vector and \( t \) is the target vector, and \( C \) is the total number of classes. Cross-entropy is a loss function used to measure the performance of the model whose output is a probability between 0 and 1. The values of the convolution kernels are initialized with random values from normal distribution and then adjusted according to the error.
The proposed model of 1D-CNN for heartbeat classification is shown in Fig. 3. It consists of 10 layers in addition to input and output layers, the first two layers are convolutional layers with 32 filters and kernel size of three, followed by Max pooling layer with pool size of three, followed by two convolutional layers with 64 filters and kernel size of three, followed by another Max pooling layer with pool size of five and final convolutional layer with 128 filters and kernel size of three. After that, three fully connected layers are added with number of neurons 256, 128, and 64 respectively. Finally, Softmax function is used in the output layer which contains 14 neurons corresponding to the 14 classes.
4 Experimental Results
4.1 Dataset
MIT-BIH dataset [26] is the most popular dataset for the existing studies. It consists of 48 records, each one is a 30-min-long with a sampling frequency of 360 Hz.
ANSI/AAMI EC57: 1998 standard [27] recommends only 44 records that can be utilized because there are four paced records. Each record is attached with a file containing the beats annotations and the R peak locations. These given annotations and locations have been used as the ground truth in the training and the evaluation steps. ECG data from lead1 only have been considered.
In this study, the beats of the 44 records are divided into training and testing portions. The data division in [3, 25] have been followed exactly for comparison sake. The percentage of training and testing portions is not equal for all classes because the number of samples for the classes is not equally distributed. The train set of the normal class consists of 13% of the total number of beats as it contains thousands of numbers of beats. On the other hand, training percentage of 40% is considered for other classes that have lower number of beats. Finally, for the classes that have a very limited number of beats, the percentage is 50%. The division of the beats is described in Table 1.
4.2 Results
In this study, MIT-BIH records are segmented into heartbeats. Thereafter, the train set is selected randomly of 21272 beats and the other 81780 beats were used as the test set. There is no duplication between the train and test sets. After training, the CNN is utilized to classify the test data. The Adam optimizer [32] is utilized to train our deep neural network, the network weights are initialized with random distribution.
The proposed method is applied to Lead1 only from the MIT-BIH arrythmia dataset and 14 classes are considered. The evaluation is done by measuring the overall accuracy. Using the average of ten trials, the achieved overall accuracy is 0.978 after 15 epochs of training for each trial. The accuracy of the test set and the loss through each epoch are shown in Fig. 4.
The comparison between our work and the other existing studies is given in Table 2. It shows that the proposed method is applied to large number of classes and the overall accuracy has been improved compared to the published results.
5 Conclusion
To conclude, an end-to-end method has been applied to classify different heartbeats into 14 classes of MIT-BIH arrythmia dataset. Dynamic heartbeat segmentation method is used to be invariant to the variability of the heart rate. Overall accuracy of 97.80% is achieved using ten trials, which outperforms the existing studies as well as more classes (14 classes) are considered. This means that CNN succeeded to learn the most important features automatically without any hand-engineering. Since this study deals with single lead signal (Lead1), further research work will be done to deal with the two channels. Furthermore, different deep learning techniques will be utilized to improve the accuracy in addition to measuring the performance of the models with more metrics such as precision and recall.
References
Tantawi, M., Revett, K., Salem, A.-B., Tolba, M.F.: Electrocardiogram (ECG): a new burgeoning utility for biometric recognition. In: Hassanien, A., Kim, T.H., Kacprzyk, J., Awad, A. (eds.) Bio-Inspiring Cyber Security and Cloud Services: Trends and Innovations, Intelligent Systems Reference Library, vol. 70, pp. 349–382. Springer, Heidelberg (2014)
Artis, S.G., Mark, R.G., Moody, G.B.: Detection of atrial fibrillation using artificial neural networks. In: Proceedings of the Computers in Cardiology, Venice, Italy, 23–26 September 1991, IEEE, Piscataway, NJ, USA, pp. 173–176 (1991)
Ye, C., Kumar, B.V.K.V., Coimbra, M.T.: Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. 59(10), 2930–2941 (2012)
Martis, R.J., Acharya, U.R., Min, L.C.: ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 8(5), 437–448 (2013)
Alfonso, V.X., Tompkins, J.: Detecting ventricular fibrillation. IEEE Trans. Biomed. Eng. 54(1), 174–177 (2007)
Minami, K., Nakajima, H., Toyoshima, T.: Real-time discrimination of ventricular tachyarrhythmia with Fouriertransform neural network. IEEE Trans. Biomed. Eng. 46(2), 179–185 (1999)
Ye, C., Vijaya Kumar, B.V.K., Coimbra, M.T.: Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. (2012)
Kastor, J.A.: Arrhythmias, 2nd edn. W.B. Saunders, London (1994)
Moody, G.B., Mark, R.G.: The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 20, 45–50 (2001). [CrossRef] [PubMed]
El-Saadawy, H., Tantawi, M., Shedeed, H.A., Tolba, M.F.: Electrocardiogram (ECG) classification based on dynamic beats segmentation. In: Proceedings of the 10th International Conference on Informatics and Systems - INFOS 2016 (2016). https://doi.org/10.1145/2908446.2908452
Gautam, M.K., Giri, V.K.: A neural network approach and wavelet analysis for ECG classification. In: Proceedings of the 2016 IEEE International Conference on Engineering and Technology (ICETECH), Coimbatore, India, 17–18 March 2016, IEEE: Piscataway, NJ, USA, pp. 1136–1141 (2016)
Zebardast, B., Ghaffari, A., Masdari, M.: A new generalized regression artificial neural networks approach for diagnosing heart disease. Int. J. Innov. Appl. Stud. 4, 679 (2013)
Min, S., Lee, B., Yoon, S.: Deep learning in bioinformatics. Briefings Bioinform. 18, 851–869 (2017)
Bakator, M., Radosav, D.: Deep learning and medical diagnosis: a review of literature. Multimodal Technol. Interact. 2, 47 (2018). https://doi.org/10.3390/mti2030047
Bengio, Y.: Learning deep architectures for AI. Found. Trends Mach. Learn. 2, 1–127 (2009)
Yu, S.N., Chou, K.T.: Integration of independent component analysis and neural networks for ECG beat classification. Expert Syst. Appl. 34, 2841–2846 (2008)
Sahoo, S., Kanungo, B., Behera, S., Sabut, S.: Multiresolution wavelet transform based feature extraction and ECG classification to detect cardiac abnormalities. Measurement 108, 55–66 (2017)
Martis, R.J., Acharya, U.R., Min, L.C.: ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 8, 437–448 (2013)
Martis, R.J., Acharya, U.R., Mandana, K., Ray, A.K., Chakraborty, C.: Application of principal component analysis to ECG signals for automated diagnosis of cardiac health. Expert Syst. Appl. 39, 11792–11800 (2012)
Yazdanian, H., Nomani, A., Yazdchi, M.R.: Autonomous detection of heartbeats and categorizing them by using support vector machines. IEEE (2013)
Elhaj, F.A., Salim, N., Harris, A.R., Swee, T.T., Ahmed, T.: Arrhythmia recognition and classification using combined linear and nonlinear features of ECG signals. Comput. Meth. Programs Biomed. 127, 52–63 (2016)
Osowski, S., Hoai, L.T., Markiewicz, T.: Support vector machine-based expert system for reliable heartbeat recognition. IEEE Trans. Biomed. Eng. 51, 582–589 (2004)
Khazaee, A.: Combining SVM and PSO for PVC detection. Int. J. Adv. Eng. Sci. 3(4) (2013)
Khalaf, A.F., Owis, M.I., Yassine, I.A.: A novel technique for cardiac arrhythmia classification using spectral correlation and support vector machines. Expert Syst. Appl. 42, 8361–8368 (2015)
El-Saadawy, H., Tantawi, M., Shedeed, H.A., Tolba, M.F.: Hybrid hierarchical method for electrocardiogram heartbeat classification. IET Signal Process. 12(4), 506–513 (2018). https://doi.org/10.1049/iet-spr.2017.0108
MIT-BIH Arrhythmias Database. http://www.physionet.org/physiobank/database/mitdb/. Accessed 12 Oct 2017
Testing and reporting performance results of cardiac rhythm and ST segment measurement algorithms. ANSI/AAMI EC57:1998 standard, Association for the Advancement of Medical Instrumentation (1998)
Thakor, N.V., Webster, J.G., Tompkins, W.J.: Estimation of QRS Complex Power Spectra for Design of a QRS Filter. IEEE Trans. Biomed. Eng. BME-31(11), 702–706 (1984). https://doi.org/10.1109/tbme.1984.325393
Yang, W., Si, Y., Wang, D., Guo, B.: Automatic recognition of arrhythmia based on principal component analysis network and linear support vector machine. Comput. Biol. Med. 101, 22–32 (2018)
Acharya, U.R., Oh, S.L., Hagiwara, Y., Tan, J.H., Adam, M., Gertych, A., Tan, R.S.: A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 89, 389–396 (2017)
Yildirim, Ö.: A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput. Biol. Med. 96, 189–202 (2018). https://doi.org/10.1016/j.compbiomed.2018.03.016
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. CoRR, abs/1412.6980 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Shaker, A.M., Tantawi, M., Shedeed, H.A., Tolba, M.F. (2020). Heartbeat Classification Using 1D Convolutional Neural Networks. In: Hassanien, A., Shaalan, K., Tolba, M. (eds) Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2019. AISI 2019. Advances in Intelligent Systems and Computing, vol 1058. Springer, Cham. https://doi.org/10.1007/978-3-030-31129-2_46
Download citation
DOI: https://doi.org/10.1007/978-3-030-31129-2_46
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31128-5
Online ISBN: 978-3-030-31129-2
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)