Abstract
There has been an increasing demand for computer-aided diagnosis systems to become self-explainable. However, in fields such as dermoscopy image analysis this comes at the cost of asking physicians to annotate datasets in a detailed way, such that they simultaneously identify and manually segment regions of medical interest (dermoscopic criteria) in the images. The segmentations are then used to train an automatic detection system to reproduce the procedure. Unfortunately, providing manual segmentations is a cumbersome and time consuming task that may not be generalized to large amounts of data. Thus, this work aims to understand how much information is really needed for a system to learn to detect dermoscopic criteria. In particular, we will show that given sufficient data, it is possible to train a model to detect dermoscopic criteria solely using global annotations at the image level, and achieve similar performances to that of a fully supervised approach, where the model has access to local annotations at the pixel level (segmentations).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Annual reports, such as [13], show that the incidence rates of skin cancer, in particular of its most aggressive form (melanoma), have been steadily increasing for the past decades. Although dermoscopy has been shown to be a powerful imaging technique, the diagnosis of dermoscopy images remains subjective a challenge for untrained dermatologists [2]. This has favored the development of computer-aided diagnosis (CAD) systems that can perform a preliminary screening of the dermoscopy images and work as second opinion tool, from which inexperienced doctors may learn [11, 12].
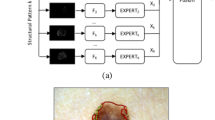
One of the most challenging aspects of CAD systems is the selection of discriminative features to characterize the dermoscopy images [4]. Earlier works based their image descriptors on the medical ABCD rule, which amounted to compute abstract asymmetry, border, color, and texture descriptors, while most recent methods avoid the process of feature design and rely on deep learning architectures to learn the most discriminative features. In both cases, the features lack medical meaning, making it hard to: (i) understand the diagnosis and (ii) be accepted by the medical community. This issue has fostered a promising line of research, where the goal is to develop clinically oriented systems that extract features that have a medical meaning, namely dermoscopic structures and colors [8]. However, this is a challenging field, due to the subtlety of several of the dermoscopic criteria and the need for extensively annotated datasets. These datasets must not only comprise the diagnosis of a skin lesion, but also detailed information of the observed dermoscopic criteria, such as segmentations at the pixel level, which we will refer to as local annotations. Until very recently, a dataset of this kind was not publicly available, which fomented the proposal of weakly supervised methods [3, 10] that could deal with the only available information (global annotations, i.e., labels at the image level), and still be able to localize the criteria in the skin lesions. Figure 1, shows the difference between local and global annotations.
Very recently, the International Skin Imaging Collaboration (ISIC), released a dataset that contained more than 2000 images, each with local annotations for a small set of relevant dermoscopic structures (pigment network, streaks, milia-like cysts, and negative network) [9]. This dataset has the potential to push forward the development of clinically inspired systems. Nonetheless, several dermoscopic criteria are still missing and the number of examples for most of the structures is small.
For dermatologists to provide local annotations is a cumbersome and time consuming task. Thus, it is important to understand if this information is really needed or if it is possible to develop a clinically inspired method, solely using global labels. This paper addresses the aforementioned issue and performs a comparison between a fully supervised system, which learns to detect dermoscopic criteria from local annotations, and a weakly-supervised system that only uses global annotations to perform the same task.
2 Proposed Framework
The goal of this work is to compare the performance of a supervised learning system against a weakly supervised one on the task of localizing four different dermoscopic structures (pigment network, negative network, streaks, and milia-like cysts) in skin lesions. The block diagram for training and testing the methods is shown in Figs. 1 and 2. The first diagram clearly states the main different between the supervised and weakly supervised strategies: the first relies on labels at the pixel level, called local annotations, thus has access to very detailed information, while the latter relies on less informative text labels at the image level (global annotations). The second diagram shows that we expect to obtain the same output with both methods, i.e., a set of binary masks with the location of the structures.
Below, we succinctly describe the steps that are common to the two methods. The supervised method is detailed in Sect. 3, while the weakly supervised approach is described in Sect. 4.
2.1 Lesion Segmentation and Superpixel Extraction
Skin lesions are segmented in order to separate them from the surrounding healthy skin. In this work we use manual segmentation masks performed by experts [9].
The following step is to divide the lesion into small and homogeneous regions, called superpixels, such that each region can be analyzed independently and classified regarding the presence/absence of each dermoscopic structure. The algorithm used to compute the superpixels is SLIC0 [1, 9].
2.2 Superpixel Characterization
The superpixel algorithm leads to the segmentation of the skin lesion into N regions that must be characterized by a feature vector \(r_{n} \in \mathbb {R} ^{f }\), such that an image d may be represented by the following set \(\mathbf r ^{d} = \{r_{1}^{d},...,r_{N}^{d}\} \in \mathbb {R} ^{f\times N^{d}}\), where \(N^{d}\) is the number of superpixels of that image. We rely on color and texture information to characterize the superpixels, in particular:
Color features: This property is characterized using the mean color vector in the HSV space (\(\mu _{\text {HSV}}\)).
Texture features: Three types of descriptors are used to characterize the texture of a region. The mean contrast (\(\mu _{\text {c}}\)), the mean contrast \(\times \) anisotropy (\(\mu _{\text {ca}}\)), and statistics computed using the directional filters from [5]. Both contrast and anisotropy are computed from the second moment matrix estimated at each pixel M(x, y)
where \(\lambda _{1}\), \(\lambda _{2}\) are the eigenvalues of M(x, y) [7]. The directional filters are applied at different orientations \(\theta _{i} \in [0,\pi ]\), \(i=0,\dots ,9\), with the impulse response for any direction \(\theta _{i}\) given by
where \(G_k\) is a Gaussian filter:
\(C_{k}\) is a normalization constant and the values of \((x',y')\) are related with (x, y) by a rotation of amplitude \(\theta _{i}\).
We compute the output of the directional filters (2) for all the directions and keep the maximum and minimum at each pixel (x, y). The regions are described by the mean and standard deviation of these values (\(\mu _{M}\), \(\sigma _{M}\), \(\mu _{m}\), and \(\sigma _{m}\)).
3 Supervised Model
In the supervised context, the model has access to all detailed information during the training phase. In the case of this work, this means that to be able to train a supervised model, we need to have access to detailed ground-truth annotations at the pixel or superpixel level (local annotations, as shown in Fig. 3). The annotations used in this work are at the superpixel level.
Given the local annotations, the problem of localizing dermoscopic structures becomes a simple classification problem, where our goal is to classify each superpixel into one of four possible classes: negative network, pigment network, milia-like cysts, and streaks. Although this could be treated as a multi-class problem, some of the superpixels have more than one label (see Fig. 3). Thus, we will train a separate classifier for each of the classes, as shown in Fig. 1. During the test phase, each of the classifiers is separately used to label the superpixels.
The classification algorithm used in this work is SVM (support vector machines) with an radial basis function (RBF) kernel.
4 Weakly Supervised Model
The supervised approached described in Sect. 3 corresponds to the ideal scenario, where we have access to detailed annotations of the presence and location of the dermoscopic structures that we want to detected. However, until the recent release of challenge related datasets, such as ISIC 2017 [9], this kind of information was unavailable. Most of times, one has only access to the dermoscopy images and a set of global annotations at the image level, which dermoscopic structures are present. To address this limitation and still be able to localize the dermoscopic structures, Barata et al. [3] proposed a framework based on the correspondence latent Dirichlet allocation (corr-LDA) model [6].
Corr-LDA belongs to the family of generative algorithms for image captioning [6]. The main idea of this method is that we can represent an image as a distribution over a set of K latent variables z, called topics. Each of the topics allow us to simultaneously express: (i) a distribution over superpixel features  , where
, where  is the set of parameters of the distribution associate to \(z_{k}\); and (ii) a multinomial distribution over the possible global annotations \(p(w|z_{k},\beta _{k})\) with parameter \(\beta _{k}\). Thus, through the topics, we are able to find the relationship between superpixel features \(r_{n}\) and the global annotations, and consequently label each superpixel.
is the set of parameters of the distribution associate to \(z_{k}\); and (ii) a multinomial distribution over the possible global annotations \(p(w|z_{k},\beta _{k})\) with parameter \(\beta _{k}\). Thus, through the topics, we are able to find the relationship between superpixel features \(r_{n}\) and the global annotations, and consequently label each superpixel.
Training the corr-LDA model amounts to estimating the set of parameters  , given a set of superpixel features for different images \(\mathcal {R} = \{\mathbf {r}^{1},\dots ,\mathbf {r}^{D}\}\) and their corresponding global annotations (negative network, pigment network, milia-like cysts, and streaks). We use the strategy described in [3, 6], to train our model. On the test phase, we apply the estimated model and compute the probability \(p(w|r_{n})\) (see [3]), to determine the probability of each global label. Since some of the superpixels may be associated with more than one label (recall Fig. 3), we experimentally determined a threshold on the probability of each annotation, such that a dermoscopic structure was assigned to a superpixel if its probability was greater than the threshold.
, given a set of superpixel features for different images \(\mathcal {R} = \{\mathbf {r}^{1},\dots ,\mathbf {r}^{D}\}\) and their corresponding global annotations (negative network, pigment network, milia-like cysts, and streaks). We use the strategy described in [3, 6], to train our model. On the test phase, we apply the estimated model and compute the probability \(p(w|r_{n})\) (see [3]), to determine the probability of each global label. Since some of the superpixels may be associated with more than one label (recall Fig. 3), we experimentally determined a threshold on the probability of each annotation, such that a dermoscopic structure was assigned to a superpixel if its probability was greater than the threshold.
5 Experimental Results
5.1 Dataset and Performance Metrics
The supervised and weakly supervised approaches were evaluated using the ISIC 2017 challenge dataset [9], which comprises 2750 images divided into training, validation, and test sets. The training set was used to estimate parameters of the supervised (SVM) and weakly supervised (corr-LDA) models, while the validation set was used to selected the best model and respective hyperparameters (the kernel width in RBF and the number of topics K in corr-LDA). The test set was used to evaluate and compare the models.
Each of the images was segmented by experts, who also annotated each of the superpixels into one or more of the following dermoscopic structures: negative network, pigment network, milia-like cysts, and streaks. Table 1 shows the number of superpixels per set, as well as the percentage that is associated to each structure. Since the weakly supervised model uses image-level annotations, we have defined that an image receives the global annotation of a specific structure if it has at least one superpixel with that label. For computational reasons it was necessary to define an additional global annotation called “without structure” to deal with images that do not exhibit any structure. The proportion of global labels per type of structure is also shown in Table 1.
These statistics show that there is a significant imbalance in the dataset, with pigment network being the most common annotation both at the superpixel and image level. To overcome this limitation, we used two strategies: (i) in the supervised approach, we have assigned different weights to the classes during the training of the classifiers, based on their distribution; (ii) in the weakly supervised approach we have artificially augmented the data, such that there were at last 500 images for each type of global annotation.
The metrics used to evaluate the models are the sensitivity (SE), specificity (SP), and balanced accuracy (BACC)
where TN, TP, FN, and FP are the total number of superpixels that are respectively true negatives, true positives, false negatives, and false positives.
5.2 Results
The scores for the supervised and weakly supervised models are shown in Table 2. In the case of the supervised model, the classification of the superpixels w.r.t to each dermoscopic structure is achieved with different degrees of success. In particular, it seems that milia-like cysts and negative network are harder to identify than the other two structures.
The weakly supervised model exhibits low SE for milia-like cysts, negative network, and streaks. However, the most noteworthy result is that of pigment network. The performance for this structure is similar to that of the supervised approach at the cost of approximately 0.43% for the annotations: recall that the supervised method relies on more than 460K local annotations at the superpixel level, while the weakly supervised uses 2K global annotations at the image level. This suggests that given a sufficient number of images with a specific dermoscopic structure, one does not require very detailed annotations as a weakly supervised model seems to be able to achieve a similar performance. Moreover, the adopted region features may also play a role, as pigment network is a highly directional structure with sharp transitions between the lines and the background. This properties may be well described by the directional filters. The performance for streaks, a structure that shares properties with pigment network, seems to support this claim, as with only 5.8% of the images with this annotation, corr-LDA still achieves a \(SE = 25.7\%\) and a \(SP = 94.5\%\) and the supervised model is the second best. Since providing detailed annotations is a cumbersome task, this result opens new possibilities for training models with less information.
Negative network and especially milia-like cysts seem to be the structures that achieve the worse performances in both supervised and weakly-supervised approaches. This suggests that the features used to characterize the superpixels may not efficiently represent these two structures. Thus, a future direction of improvement will be to replace the features described in Sect. 2.2 with more powerful descriptors, namely those based on deep neural networks.
6 Conclusions
This paper compares two models for the detection and localization of four dermoscopic structures (pigment network, milia-like cysts, negative network, and streaks) in superpixels of dermoscopy images. The first model was fully supervised, thus was trained using more than 4.60 \(\times \,10^5\) local annotations at the superpixel level, while the second model relied on a weakly-supervised algorithm (corr-LDA), trained using 2 \(\times \,10^3\) global annotations at the image level. The experimental results surprisingly showed that in the case of pigment network, the weakly supervised model is able to achieve a similar performance to that of the supervised one, with a significantly smaller amount of ground truth information. However, the supervised model achieved better performances for the remaining structures, suggesting that better features and more data is required. Nonetheless, the most relevant output of this work is that given a sufficient number of images, we may not need detailed information to be able to detect dermoscopic structures.
References
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., Susstrunk, S.: SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 34(11), 2274–2282 (2012)
Argenziano, G., Soyer, H.P., De Giorgi, V., et al.: Interactive Atlas of Dermoscopy. EDRA Medical Publishing & New Media (2000)
Barata, C., Celebi, M.E., Marques, J.S.: Development of a clinically oriented system for melanoma diagnosis. Pattern Recogn. 69, 270–285 (2017)
Barata, C., Celebi, M.E., Marques, J.S.: A survey of feature extraction in dermoscopy image analysis of skin cancer. IEEE J. Biomed. Health Inform. 23(3), 1096–1109 (2018)
Barata, C., Marques, J.S., Rozeira, J.: A system for the detection of pigment network in dermoscopy images using directional filters. IEEE Trans. Biomed. Eng. 59(10), 2744–2754 (2012)
Blei, D., Jordan, M.: Modeling annotated data. In: 26th Annual International ACM SIGIR Conference on Research and Development in Informataion Retrieval, pp. 127–134. ACM (2003)
Carson, C., Belongie, S., Greenspan, H., Malik, J.: Blobworld: image segmentation using expectation-maximization and its application to image querying. IEEE Trans. Pattern Anal. Mach. Intell. 24(8), 1026–1038 (2002)
Celebi, M.E., Codella, N., Halpern, A.: Dermoscopy image analysis: overview and future directions. IEEE J. Biomed. Health Inform. 23(2), 474–478 (2019)
Codella, N.C.F., Gutman, D., Celebi, M.E., et al.: Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In: IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 168–172 (2018)
Madooei, A., Drew, M.S., Hajimirsadeghi, H.: Learning to detect blue-white structures in dermoscopy images with weak supervision. IEEE J. Biomed. Health Inform. 23(2), 779–786 (2018)
Oliveira, R., Papa, J., Pereira, A., Tavares, J.: Computational methods for pigmented skin lesion classification in images: review and future trends. Neural Comput. Appl. 29(3), 1–24 (2016). https://doi.org/10.1007/s00521-016-2482-6
Pathan, S., Prabhu, K.G.S., Siddalingaswamy, P.C.: Techniques and algorithms for computer aided diagnosis of pigmented skin lesions - a review. Biomed. Signal Process. Control 39, 237–262 (2018)
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics, 2019. CA: Cancer J. Clin. 69, 7–34 (2019)
Acknowledgments
This work was supported by the FCT project and plurianual funding: [PTDC/EEI PRO/0426/2014], [UID/EEA/50009/2019].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ferreira, B., Barata, C., S. Marques, J. (2019). What Is the Role of Annotations in the Detection of Dermoscopic Structures?. In: Morales, A., Fierrez, J., Sánchez, J., Ribeiro, B. (eds) Pattern Recognition and Image Analysis. IbPRIA 2019. Lecture Notes in Computer Science(), vol 11868. Springer, Cham. https://doi.org/10.1007/978-3-030-31321-0_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-31321-0_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31320-3
Online ISBN: 978-3-030-31321-0
eBook Packages: Computer ScienceComputer Science (R0)