Abstract
Considering the DenseFuse only works in a single scale, we propose a multi-scale DenseNet (MSDNet) for medical image fusion. The main architecture of network is constructed by encoding network, fusion layer and decoding network. To utilize features at different scales, we add a multi-scale mechanism which uses three filters of different sizes to extract features in encoding network. More image details are obtained by increasing the encoding network’s width. Then, we adopt fusion strategy to fuse features of different scales respectively. Finally, the fused image is reconstructed by decoding network. Compared with the existing methods, the proposed method can achieve state-of-the-art fusion performance in objective and subjective assessment.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Medical image fusion plays an important role in medical clinical applications. Recently, medical image fusion mainly concentrates on computerized tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET) and single-photon emission computed tomography (SPECT) modalities [1]. Different imaging mechanism leads to different focus on medical images. For example, the CT has an advantage of dense resolution of denses structures like bones and implants, the MRI is good at soft-tissue details with high-resolution anatomical information, while the blood flow and metabolic changes can be supported by PET and SPECT images but with low spatial resolution [2]. Therefore, how to extract the salient features of different modalities and how to choose proper fusion strategy are main issues in medical image fusion.
Generally speaking, the most common method is multi-scale transform (MST) in image fusion. MST-based methods consist of decomposition, fusion and reconstruction. There are many MST-based methods for image fusion, such as discrete wavelet transform (DWT) [4], contourlet transform [3] and curvelet transform (CVT) [5] etc. Du et al. [17] presented a multiscale decomposition method based on local Laplacian filtering (LLF) with an information of interest (IOI)-based strategy for medical image fusion.
Representation learning [8, 23, 24] is widely used in image fusion in recent years. In sparse representation, Liu et al. [6] applied adaptive sparse presentation in image fusion and denoising. Yin et al. [7] proposed a novel multi-focus image fusion approach based on sparse representation which used joint dictionary. In low-rank representation (LRR), Li et al. [8] proposed a novel multi-focus image fusion based on dictionary learning and LRR to get a better performance.
With the development of deep learning, many image fusion methods based on deep learning are proposed. Because of the depth of network, we can use many deep features to make fusion. In [9], authors presented a new method based on a deep convolutional neural network (CNN). The two source images are fed to network and the score map will be obtained. In [10], Li et al. considered the features of middle layers of pretrained VGG-network [11] and used \(l_1\)-norm and weighted-average strategy to generate several candidates of the fused detail content.
In 2017, Prabhakar et al. [21] proposed an image fusion framework which consists of feature extraction layers, a fusion layer and reconstruction layers. Their feature extraction layers have Siamese network architecture and reconstruction layers consist of three CNN layers. In 2019, Li et al. [12] proposed a DenseFuse whose encoding network is combined with convolutional layers and dense block for infrared and visible images. Although this method achieves better performance, but it still has a drawback because it just considers a single scale to extract image feature in encoding network.
To solve this problem, in this paper, we improve the DenseFuse [12] with a multi-scale mechanism in encoding network. Then, we apply the improved architecture to the medical image fusion. We use three filters of different sizes to extract features of the end of original encoder of DenseFuse respectively. So that we can obtain more features maps of different scales, then, we adopt fusion strategy to fuse them respectively. Finally, we cascade the fused features of different scales into decoder to obtain the fused image.
The structure of the rest paper is organized as follows. In Sect. 2, we will briefly introduce the related work. In Sect. 3, the improved method will be presented in detail. Section 4 shows the experimental results. Finally, Sect. 5 is the conclusion of our paper.
2 Related Works
Recently, many deep learning methods are adopted in the field of image fusion. With the development of deep networks, some issues have arisen, such as the disappearance of gradients and the increase of parameters etc. In CVPR 2017, Huang et al. [13] presented the Dense Convolutional Network (DenseNet) in which the feature-maps of each layer are treated as input into all subsequent layers. This architecture has several advantages: they can make vanishing-gradient problem alleviated, make full use of the features of the middle layers and reduce the number of parameters.
Based on the advantages of DenseNet, In 2019, Li et al. [12] proposed a novel deep learning architecture, which consists of encoding network and decoding network. Their encoding network is constructed by convolutional layers and dense block. Then, the authors fuse them by fusion layer. Finally, the fused image is reconstructed by a decoder. In encoder, they apply dense block [13] to leverage more useful information from middle layers. A unique training strategy was developed in DenseFuse. Then, the output image is reconstructed by decoding network using the extracted features. Therefore, in the fusion phase, the two source images are fed to encoding network. Then, two feature maps are obtained and they are fused by fusion strategy. Ultimately, the fusion maps are transmitted to decoder network and the fused image can be obtained.
In this paper, we propose a multi-scale DenseNet (MSDNet), which is built on DenseFuse. We add multi-scale mechanism in encoder to extract features from different scales. Then, we apply fusion strategy to fuse them respectively. We utilize the decoder of Densefuse which is four CNN layers to reconstructed the fused image. We will introduce the algorithm in detail in Sect. 3.
3 Methodology
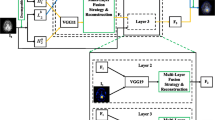
In this section, the improved method will be introduced in detail. First of all, we apply our method to medical image fusion. The framework of the method is shown in Fig. 1.
The input medical images are denoted as \(I_1\) and \(I_2\). We adopt the method [18] to convert the color images into YUV color space. If \(I_1\) is grayscale, we convert \(I_2\) to YUV space but we just leverage the Y channels of \(I_2\). We then input the two (converted) images into MSDNet. Finally, combining the output of MSDNet with U and V channels of \(I_2\), the fused image (f) is obtained by converting YUV to RGB color space.
Secondly, considering only a single scale feature is used in DenseFuse, our goal is adding multi-scale mechanism in encoder of DenseFuse. Therefore, our new framework is called MSDNet. MSDNet is made of encoder, a fusion layer and decoder as shown in Fig. 2.
The encoder is constructed by a convolutional layer, dense block and a multi-scale layer. We add multi-scale layer whose filters’ size are \(5\times 5\), \(3\times 3\) and \(1\times 1\) respectively to extract features from coarse to fine at the end of Denseblock. Why we choose these sizes of filters? Because we adopt \(1\times 1\) filters to fuse the information of different channels at the same location and \(3\times 3\), \(5\times 5\) filters to fuse the information of different channels around the same location. When we choose a larger size of filters, the features extracted from the filters will not be obvious, so larger size of filters is not selected. Through the multi-scale layer, we will get three groups of multi-channel features. Then, we choose \(l_1\)-norm strategy [12], which is shown in Fig. 3, to fuse them respectively.
In Fig. 3, the feature maps are denoted as \(\varphi _i^{1:n}(x,y)\), where \(i \in \{1,\dots , k\}\) is the number of input images, \(n \in \{1,2,\dots ,N\}\), and \(N=64\) is the number of feature maps. \(\varphi _i^{1:n}(x,y)\) are processed by \(l_1\)-norm as Eq. 1.
Then the average operator is utilized to calculate the weight map by Eq. 2.
Finally, \(f_k^n(x,y)\), where \(k \in \{1, 3, 5\}\) is the scale of filters, is calculated by Eq. 3.
The three groups of fused features \(f_1\), \(f_3\) and \(f_5\) will be concatenated together as input to decoder, lastly, we will get the reconstructed image which is the fused image.
In training phase, our aim is to train the network’s ability to reconstruct the input image, as shown in Fig. 4. The input image is extracted by dense convolutional layers, then, the convolutional kernels of different size are used to extract the features of different scales which are \(1\times 1\), \(3\times 3\) and \(5\times 5\) respectively. Finally, we concatenate the multi-channel features of different scales into the decoder. We adopt structural similarity (SSIM) loss [12] and pixel loss [12] to guarantee the reconstructed image will be closely to the input image, as Eq. 4.
In this paper, \(\lambda = 1000\), the reason will be introduced in details in Sect. 4.2.
Our training images are from MS-COCO [14]. We choose 80000 images, which are resized to \(256\times 256\) and transformed to gray scale images, are utilized to train the network. Learning rate, batch size and epochs are set as \(1\times 10^{-4}\), 2 and 4 respectively.
4 Experiments and Analysis
4.1 Experiment Settings
In our experiment, there are three fusion categories of medical images, which are computerized tomography (CT) and magnetic resonance imaging (MRI), MRI and positron emission tomography (PET), and MRI and single-photon emission computed tomography (SPECT) [20].
As shown in Fig. 1, in CT and MRI, CT is \(I_1\) and MRI is \(I_2\); in MRI and PET, MRI is \(I_1\) and PET is \(I_2\); in MRI and SPECT, MRI is \(I_1\) and SPECT is \(I_2\). Therefore, we fuse three groups of medical images and analyze them from objective and subjective points of views.
We compare the proposed method with seven prior methods, including a medical image fusion method based on convolutional neural networks (CNN) [2], IHS-PCA method [15] which adopted intensity-hue-saturation (IHS) transform and principal component analysis (PCA) to preserve more spatial feature and more required functional information with no color distortion, LES-DC [16], LLF-IOI [17], medical image fusion with PA-PCNN in nonsubsampled shearlet transform domain (NSST) [18], infrared and visible image fusion using a deep learning framework (VGG) [10], DenseFuse [12].
In order to evaluate our proposed method, we compare our method with seven existing methods and we choose six quality indicators. They are: \(SSIM_a\); \(PSNR_a\); \(FMI_{dct}\), \(FMI_w\), \(FMI_{edge}\) and \(FMI_{gradient}\) [19] which calculate mutual information for the discrete cosine, wavelet, edge and gradient features, respectively.
In our experiment, the \(SSIM_a\) and \(PSNR_a\) are calculated by Eqs. 5 and 6,
where \(SSIM(\cdot )\) denotes the structural similarity operation [22], \(PSNR(\cdot )\) denotes peak signal-to-noise ratio, F is the fused image and \(I_1\),\(I_2\) are source images. The values of \(SSIM_a\) and \(PSNR_a\) represent the ability to retain structural information and original information of source images, respectively.
With the increase of all these six measures, the image fusion performance will be improved.
We use Python for all experiments and adopt Tensorflow architecture. Our method was implemented with NVIDIA GTX 1080Ti GPU.
4.2 Loss of Training Phase
In [12], \(\lambda \in \{1, 10, 100, 1000\}\), according to the experimental comparison as shown in Fig. 5, we find that the model converges faster and more stable when \(\lambda =1000\). Therefore, in this paper, we choose \(\lambda =1000\).
4.3 Baseline
Firstly, we compare the method with DenseFuse (\(\lambda =1000\)), which is a recently developed fusion method, separately. In Table 1, the values are the average results for 60 fused images, including CT and MRI, MRI and PET and MRI and SPECT. The best results are bloded. We can see that it is effective for adding multi-scale mechanism in DenseFuse.
4.4 Subjective Evaluation
The fused images which are obtained by the seven compared methods and proposed method are shown in Fig. 6.
As we can see from Fig. 6, LES-DC [16] and VGG [10] retain less valid information than other methods because some features are not very clear. The fused images obtained by LLF-IOI [17] are a little sharp and have some artificial noise. Compared with the existing methods, the fused images obtained by our method are more natural. We will leverage objective indicators to analyze the fusion performance.
Fused results for medical images (RGB) images. Rows 1 to 3 of (a) and (b) are CT and MRI images; Rows 4 to 6 of (a) and (b) are MRI and PET images; Rows 7 to 9 of (a) and (b) are MRI and SPECT images; (c) CNN; (d) IHS-PCA; (e) LES-DC; (f) LLF-IOI; (g) NSST with PAPCNN; (h) VGG; (i) DenseFuse; (j) Ours
4.5 Objective Evaluation
We use SSIM, PSNR, \(FMI_{dct}\), \(FMI_w\), \(FMI_{edge}\), \(FMI_{gradient}\) to analyze fusion performance. We test three groups of medical images, they are: CT and MRT, MRI and PET and MRI and SPECT. The results are shown in Table 2.
In Table 2, the best results are bloded, the second-best results are marked in red and the third-best results are marked in blue. In Table 2, it can be seen that proposed method’s indicators are very high for the most part. It proves that the results of proposed method has more salient features and less artificial noise.
5 Conclusion
In this paper, we propose a multi-scale DenseNet by adding multi-scale mechanism into DenseFuse and apply the improved method for medical image fusion.
Our network consists of constructed by encoder, fusion layer and decoder. Encoder is made of a convolutional layer, dense block and multi-scale layer. Decoder is made of four CNN layers. After multi-scale layer, we obtain three groups of feature maps. Then, we utilize \(L_1\)-norm fusion strategy to fusion them respectively. We concatenate them as input into decoder. Finally, we obtain the fused image reconstructed by decoder.
We use subjective and objective quality metrics to evaluate the performance of fusion results. The results of experiments indicate that our method is effective for medical image fusion.
References
Du, J., Li, W., Lu, K., et al.: An overview of multi-modal medical image fusion. Neurocomputing 215, 3–20 (2016)
Liu, Y., Chen, X., Cheng, J., et al.: A medical image fusion method based on convolutional neural networks. In: 2017 20th International Conference on Information Fusion (Fusion), pp. 1–7. IEEE (2017)
Yang, S., Wang, M., Jiao, L., et al.: Image fusion based on a new contourlet packet. Inf. Fusion 11(2), 78–84 (2010)
Li, H., Manjunath, B.S., Mitra, S.K.: Multisensor image fusion using the wavelet transform. Graph. Models Image Process. 57(3), 235–245 (1995)
Guo, L., Dai, M., Zhu, M.: Multifocus color image fusion based on quaternion curvelet transform. Opt. Express 20(17), 18846–18860 (2012)
Liu, Y., Wang, Z.: Simultaneous image fusion and denoising with adaptive sparse representation. IET Image Proc. 9(5), 347–357 (2014)
Yin, H., Li, Y., Chai, Y., et al.: A novel sparse-representation-based multi-focus image fusion approach. Neurocomputing 216, 216–229 (2016)
Li, H., Wu, X.-J.: Multi-focus image fusion using dictionary learning and low-rank representation. In: Zhao, Y., Kong, X., Taubman, D. (eds.) ICIG 2017. LNCS, vol. 10666, pp. 675–686. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-71607-7_59
Liu, Y., Chen, X., Peng, H., et al.: Multi-focus image fusion with a deep convolutional neural network. Inf. Fusion 36, 191–207 (2017)
Li, H., Wu, X.J., Kittler, J.: Infrared and visible image fusion using a deep learning framework. In: 2018 24th International Conference on Pattern Recognition (ICPR), pp. 2705–2710. IEEE (2018)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Li, H., Wu, X.J.: DenseFuse: a fusion approach to infrared and visible images. IEEE Trans. Image Process. 28(5), 2614–2623 (2019)
Huang, G., Liu, Z., Van Der Maaten, L., et al.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Lin, T.-Y., et al.: Microsoft COCO: common objects in context. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8693, pp. 740–755. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10602-1_48
He, C., Liu, Q., Li, H., et al.: Multimodal medical image fusion based on IHS and PCA. Procedia Eng. 7, 280–285 (2010)
Xu, Z.: Medical image fusion using multi-level local extrema. Inf. Fusion 19, 38–48 (2014)
Du, J., Li, W., Xiao, B.: Anatomical-functional image fusion by information of interest in local Laplacian filtering domain. IEEE Trans. Image Process. 26(12), 5855–5866 (2017)
Yin, M., Liu, X., Liu, Y., et al.: Medical image fusion with parameter-adaptive pulse coupled neural network in nonsubsampled shearlet transform domain. IEEE Trans. Instrum. Meas. 99, 1–16 (2018)
Haghighat, M., Razian, M.A.: Fast-FMI: non-reference image fusion metric. In: 2014 IEEE 8th International Conference on Application of Information and Communication Technologies (AICT), pp. 1–3. IEEE (2014)
Prabhakar, K.R., Srikar, V.S., Babu, R.V.: DeepFuse: a deep unsupervised approach for exposure fusion with extreme exposure image pairs. In: ICCV, pp. 4724–4732 (2017)
Wang, Z., Bovik, A.C., Sheikh, H.R., et al.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13(4), 600–612 (2004)
Chen, Z., Wu, X.J., Kittler, J.: A sparse regularized nuclear norm based matrix regression for face recognition with contiguous occlusion. Pattern Recogn. Lett. (2019)
Chen, Z., Wu, X.-J., Yin, H.-F., Kittler, J.: Robust low-rank recovery with a distance-measure structure for face recognition. In: Geng, X., Kang, B.-H. (eds.) PRICAI 2018. LNCS (LNAI), vol. 11013, pp. 464–472. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-97310-4_53
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Song, X., Wu, XJ., Li, H. (2019). MSDNet for Medical Image Fusion. In: Zhao, Y., Barnes, N., Chen, B., Westermann, R., Kong, X., Lin, C. (eds) Image and Graphics. ICIG 2019. Lecture Notes in Computer Science(), vol 11902. Springer, Cham. https://doi.org/10.1007/978-3-030-34110-7_24
Download citation
DOI: https://doi.org/10.1007/978-3-030-34110-7_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34109-1
Online ISBN: 978-3-030-34110-7
eBook Packages: Computer ScienceComputer Science (R0)