Abstract
Recent deep neural networks have achieved great success in medical image segmentation. However, massive labeled training data should be provided during network training, which is time consuming with intensive labor work and even requires expertise knowledge. To address such challenge, inspired by typical GANs, we propose a novel end-to-end semi-supervised adversarial learning framework for medical image segmentation, called “Importance guided Semi-supervised Deep Networks” (ISDNet). While most existing works based on GANs use a classifier discriminator to achieve adversarial learning, we combine a fully convolutional discriminator and a classifier discriminator to fulfill better adversarial learning and self-taught learning. Specifically, we propose an importance weight network combined with our FCN-based confidence network, which can assist segmentation network to learn better local and global information. Extensive experiments are conducted on the LASC 2013 and the LiTS 2017 datasets to demonstrate the effectiveness of our approach.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Medical image segmentation is a fundamental yet challenging problem in medical image analysis, which aims to segment organs or pathological area (e.g. left atrium or liver lesion) in medical images. Recently, a typical Fully Convolutional Network (FCN) [20] based methods of encoder-decoder structure have achieved considerable success in semantic image segmentation, such as U-Net [21] and deeplabv3+ [5]. Although CNN-based approaches have made great progress, they often require massive labeled training data. Medical image tasks are usually more difficult than natural image tasks because of little training data and high cost manual annotations, etc. In addition, most medical datasets are still constrained in limited size and application scenarios. Hence, it is a complex and expensive procedure to obtain large-scale medical labeled data.
In order to ease the effort of acquiring high-quality data, unsupervised learning is an alternative way to utilize massive unlabeled data. However, due to lacking the concept of classes, unsupervised learning methods have not attained convincing performance in segmentation. So semi/weakly-supervised methods have drawn much attention from many researchers in the community. These methods generally demand additional annotations or data, such as the image-level class label [18], box level [16, 17], point level [19], or scribble level [1].
Recently, Generative Adversarial Networks (GANs) [22] have achieved a wide range of success due to their ability to generate high-quality realistic images [2, 6, 11, 12]. It attracts extensive attention with successful applications in super-resolution [14], domain adaptation [8, 15], zero-shot learning [13], etc. A classic GAN consists of two sub-networks (i.e., generator and discriminator) that play a min-max game in the training procedure where generator produces a sample of the target data distribution, while discriminator aims to differentiate between real and fake data repeatedly. The generator is then optimized to generate more realistic samples that are more close to target data distribution. Recently, several works have applied the GANs framework in semantic segmentation. Luc et al. [23] propose to apply a classifier discriminator to assist the training process for semantic segmentation in a fully-supervised way. But this method has not achieved distinguished results over the baseline scheme and fails to tackle unlabeled data for semi-supervised setting. Frid-Adar et al. [9] employ GANs to generate synthetic medical images to train segmentation network. Nie et al. [7] use sample attention mechanism to improve the network training. However, those jobs utilize the classification networks as their discriminators in fully-supervised ways, so those discriminators only can capture the global information of generated masks and ground truth masks without considering the local details, which is more important for the task of semantic segmentation.
In this paper, we propose a semi-supervised semantic segmentation algorithm (called ISDNet) based on GANs in order to alleviate the demand for large-scale medical labeled data. Inspired by [10] and [8], our network consists of three sub-networks: (1) segmentation network, (2) confidence network and (3) importance weight network. In this work, two semi-supervised loss are conducted to leverage the unlabeled data. First, Our FCN-based confidence network can generate confidence maps, which can guide our segmentation network in a self training strategy. The confidence maps provide us the trustworthy regions in the segmented label map, which can be selected to generate proxy label for unlabeled data. Second, we apply the adversarial loss on unlabeled data in the supervised setting, which encourages the model to predict segmentation outputs of unlabeled data close to the ground truth distributions. Then, our importance weight network can identify the importance score of unlabeled samples, which represents the probability of the sample come from the labeled data distribution. Finally, we integrate the importance score into semi-supervised loss and obtain our two weighted semi-supervised loss.
In sum, our main contributions include: (i) we develop an adversarial framework, which improves semantic segmentation performance without adding any computational cost during inference; (ii) we propose a semi-supervised framework to improve the segmentation accuracy with unlabeled data; (iii) we combine a fully convolutional discriminator and a classifier discriminator to facilitate the semi-supervised learning, which can better use unlabeled data. To demonstrate the effectiveness of our proposed approach, ablation studies are conducted on the LASC 2013 dataset. Overall, our proposed approach brings 1.8% improvement on the LASC 2013 dataset and 11.4% improvement on the LiTS 2017 dataset.
2 Our Approach
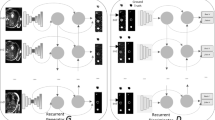
The proposed ISDNet consists of three subnetworks, i.e., (1) segmentation network (denoted as S), (2) confidence network (denoted as D) and (3) importance weight network (denoted as \(D_0\)). The architecture of our proposed framework is presented in Fig. 1.
In order to facilitate elaboration, we first give the definitions of terminologies used throughout the paper. Given an input medical image \(X_n\) of dimension \({H\times W\times 1}\) and its one-hot encoded ground truth label \(Y_n\), we denote the segmentation network as \(S(\cdot )\), the features as \(Z(X_n)\) extracted from the encoder of our segmentation network and the predicted probability map as \(\hat{Y}_n=S(X_n)\) of size \({H\times W\times C}\), where C is the category number. We denote our fully convolutional discriminator as \(D(\cdot \)) which takes class probability maps as the input (the output from segmentation network \(S(X_n)\) or ground truth label maps \(Y_n\)) and then outputs a confidence map of size \({H\times W\times 1}\). Confidence map generated by D scores each pixel p, which represents whether that pixel is sampled from the ground truth label (p = 1) or the segmentation network (p = 0). The importance weight network denoted as \(D_0(\cdot )\), takes \(Z(X_n)\) (the features of labeled sample or unlabeled sample) as the input and outputs an importance weight. The importance weight represents whether that sample is sampled from the labeled data (\({X_n =1}\)) or the unlabeled data (\({X_n=0}\)).
2.1 Segmentation Network
In ISDNet as shown in Fig. 1, we employ a simplified 2D U-Net (SU-Net) as our segmentation network, but any end-to-end segmentation network can be applied, such as FCN and deeplabv3+, etc. In this paper, we halve the number of all convolutional layers in original U-Net [21], and use leaky ReLU and group normalization [3] instead of ReLU and batch normalization to balance the performance and memory cost.
Total Loss for Segmentation Network. We train the segmentation network by minimizing a multi-task loss function
where \(L_{Dice}\), \(L_{wadv}\) and \(L_{wsemi}\) denote the multi-class dice loss, adversarial loss and semi-supervised loss, respectively. In (1), \(\lambda _{adv}\) and \(\lambda _{semi}\) are two weights for minimizing the proposed multi-task loss function.
Multi-class Dice Loss. To overcome the class imbalance problem, we propose to use a weighted multi-class dice loss as the segmentation loss
where \(\hat{Y}_n^c\) denotes the predicted probability belonging to class c (i.e. background, liver, or liver lesion), \(Y_n^c\) denotes the ground truth probability, and \(w^c\) denotes a class dependent weighting factor. Empirically, we set the weights to be 0.2 for background, 1.2 for liver, and 2.2 for liver lesion. But for left atrium (LA) segmentation, we use normal two-class dice loss.
2.2 Confidence Network
Different from using CNN-based discriminator, we propose to use a FCN-based discriminator called confidence network to generate more detailed adversarial information in local region. Hence, we combine adversarial learning in our work to further optimize the segmentation network.
Adversarial Loss of the Confidence Network. To train the confidence network, we minimize the binary cross-entropy loss \(L_{D}\) using
where \(X_n\) and \(Y_n\) represent the input data and its corresponding ground truth label map (one-hot encoding scheme), respectively. In addition, \(D(S(X_n))^{(h,w)}\) is the confidence map of \(X_n\) at location (h, w), and \(D(Y_n)^{(h,w)}\) is defined similarly.
Adversarial Loss of the Segmentation Network. In the conventional GANs, generators and discriminators play a min-max game. Hence, there is another loss from D working as adversarial Loss to further improve segmentation network. It enforces higher-order consistency between ground truth label and predicted masks. We utilize the loss \(L_{adv}\) based on a fully convolutional discriminator network
With this loss, we further improve the ability of segmentation network to fool the discriminator by maximizing the probability of the predicted results being more close to the ground truth distribution.
2.3 Importance Weight Network
Training with Unlabeled Data. In this work, we concern more about how to use unlabeled data to improve the performance of segmentation network. Since there is no corresponding ground truth label for unlabeled data, \(L_{Dice}\) no longer works. But the adversarial loss \(L_{adv}\) is still applicable as it only requires the discriminator network.
In addition, since our confidence network could provide local confidence information, we propose a self-taught learning framework with our trained discriminator D for unlabeled data. The main idea is that our confidence network can generate a confidence map \(CM=D(S(X_n))^{h,w}\) to indicate us which regions of the predicted results are sufficiently close to the ground truth distribution. Then we process the confidence map with a threshold \(T_{semi}\) to obtain the confident regions. In this way, we can exploit these confident regions to filter the segmentation results of unlabeled data to improve the segmentation network. The semi-supervised loss is defined as
where \(\bar{Y}_n\) is the one-hot encoding of \(\arg \max (\hat{Y}_n)\). \([\cdot ]\) is the indicator function.
Sample Weights Learning. Furthermore, we employ another discriminator named \(D_0\). It can output the probability that an unlabeled sample belongs to labeled data distribution on the image level. Specifically, the importance weight network is similar to the original GANs with min-max loss
where \(Z(\cdot )\) is the feature extractor (also is the encoder of our SU-Net) for labeled data \(X_l\) and unlabeled data \(X_{unl}\) respectively, and \(D_0\) is a binary classifier with all the labeled data labeled as 1 and all the unlabeled data labeled as 0.
Assume that in the case of the optimal classifier \(D_0\), the output value of \(D_0\) is the probability that the sample comes from the labeled data distribution. If \(D^{*}(z) \approx 1\), then the sample will highly likely originate from the labeled data distribution, since the features \(Z(X_l)\) are quite different from the features \(Z(X_{unl})\) and can be ideally separated from labeled data distribution by \(D_0\). Then we should reduce the contribution of these samples because feature extractor has already been trained by some similar samples. On the other hand, if \(D^{*}(z)\) is small, it means feature extractor has not been trained by those samples. A larger importance weight should be applied to these samples to improve segmentation network. Hence, the sample weights function should be inversely related to \(D^{*}(z)\) and the importance weight function of the unlabeled samples is defined as
As can be seen that if \(D^{*}(z)\) is small, w(z) is large. Hence, the weights for unlabeled samples that are similar to the labeled data will be smaller than those are not similar to the labeled data. Our aim is to obtain the relative importance of unlabeled samples. The weight function can also be expressed as a function of density ratio between labeled and unlabeled features. If we apply the weights to \(D_0\), then the Jensen-Shannon divergence between two densities can not be reduced from the theoretical results of the minimax game [8]. Hence, we utilize the weights to D to solve this issue. In this way, \(D_0\) is only used for obtaining the importance weights for unlabeled samples. Thus, we will not update the encoder with the gradient of \(D_0\). So we can integrate the importance weight into our semi-supervised loss.
And adversarial loss for unlabeled data can be modified as
But for labeled data, w(z) should be set to 1. By summing the above losses, the total loss to train the segmentation network can be defined as (1).
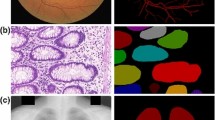
Comparisons on the LiTS 2017 dataset using 1/2 labeled data. Green area and red area represent liver and lesion, respectively. It can be seen that our confidence network assists segmentation network discover parts of lesions while our baseline does not. Furthermore, our semi-supervised algorithm refines the segmentation results.
3 Experiments
Evaluation Datasets and Metrics. Experiments are conducted on two publicly available datasets to report the state-of-the-art performance of ISDNet. Ablation studies are conducted on LASC 2013 dataset [4]. We also conduct experiments on LiTS 2017 dataset to verify the validity of our method. For LASC 2013, we employ data augmentation and obtain 3 K 2D images with size \(320\times 320\) in total. Dice index for left atrium (LA) and running time are selected to compare with other state-of-the-art method. For LiTS 2017, we use two metrics to evaluate the segmented liver lesions, including dice per case and global dice.
Implementation Details. All experiments are built with Pytorch framework on a single NVIDIA 1080ti GPU. We use the Adam optimizer for both our segmentation network and two discriminators with the learning rate \(10^{-4}\). For the hyper-parameters in the proposed method, \(\lambda _{adv}\) is set as 0.001 when trained with labeled and unlabeled data. We set \(\lambda _{semi}\) as 1 and \(T_{semi}\) as 0.1. For semi-supervised training, we randomly divide all dataset into labeled and unlabeled data. We initiate the semi-supervised learning after 5 epochs training with labeled data. In each epoch, we train both the segmentation network and two discriminator networks, only labeled data are used for training of the discriminator D while \(D_0\) demands the part of unlabeled data.
3.1 LASC 2013
Results and Analysis. Table 1 shows the evaluation results on the LASC 2013 test dataset. We randomly sample 1/2 images as labeled data, the rest as unlabeled data. We compare the proposed algorithm against LTSI-VRG [24], UCL-1C [25], UCL-4C [25], U-Net [21, 26] and [27] to demonstrate that our baseline model (SU-Net) performs comparably with the state-of-the-art schemes. LTSI-VRG, UCL-1C, UCL-4C are the top three methods in Dice index, which are all based on multi-atlas. OBS-2 [4] is the result from human observer. [26] combines CNN and recurrent neural network (RNN) to achieve a Dice index of 0.94, but it will greatly increase the inference time (22 times slower than our SU-Net). [27] is based on a multi-view CNN with an adaptive fusion strategy and a new loss function, which is 400 times slower than ours and use 20 times more data than us. Table 1 shows that the adversarial loss brings consistent performance improvement (from 0.8% to 1.0%) over different amounts of training data. Incorporating the proposed semi-supervised learning scheme brings overall 1.8% improvement. For [10] and [23], we use our SU-Net to replace its original segmentation network for equal comparisons. Specially, our importance weight network brings 0.3% improvement compared to [10]. It means our network makes better use of unlabeled data to improve network performance. Apart from this, our confidence network brings improvement (from 0.4% to 0.5%) compared to a typical classifier discriminator [23].
Hyper-parameter Analysis. The proposed algorithm is governed by three hyper parameters: \(\lambda _{adv}\) and \(\lambda _{semi}\) for balancing the multi-task learning in (1), and \(T_{semi}\) used to control the sensitivity in the semi-supervised learning described in (5). Table 2 shows sensitivity analysis of hyper parameters on the LASC 2013 dataset under semi-supervised setting. Different from [10], we find that smaller \(\lambda _{adv}\) must be used for medical image tasks, this is because the content of natural images is richer and requires larger loss to guide network learning. Then, we conduct the experiments with different values of \(T_{semi}\). With higher \(T_{semi}\), our algorithm will select regions, which are more close to the ground truth distribution. When \(T_{semi}=0\), all the pixel predictions in unlabeled images will be applied for semi-supervised training, which leads to performance degradation. Overall, the proposed model achieves the best results when \(T_{semi}=0.1\).
Ablation Study. We present ablation study of our proposed system in Table 3 on LASC test dataset. Our confidence network gains 0.5% and 0.4% improvement compared to a classifier discriminator with half and full data, respectively. Then, we apply the semi-supervised learning method without the adversarial loss. The results show that the adversarial procedure on the labeled data is necessary to our semi-supervised scheme. If the segmentation network does not participate in adversarial training, the confidence maps generated by the discriminator would be pointless. As shown in Table 3, our semi-supervised methods in ISDNet help to improve segmentation performance.
3.2 LiTS 2017
Results and Visualization. Furthermore, we extend the experiment on LiTS 2017 dataset. Figure 2 shows visual comparisons of the segmentation results on the LiTS 2017 validation dataset generated by our proposed method. It can be seen that no lesion is found in our baseline (Fig. 2(b)), but with the assistance of adversarial loss, segmentation network can detect parts of lesions (Fig. 2(c)). Further more, with our semi-supervised adversarial learning algorithm, segmentation network could segment a majority of lesions (Fig. 2(d)). Table 4 shows the liver lesion evaluation results on the LiTS 2017 test dataset with random sampled 1/2 images as labeled data. It can be seen that our methods have made the best dice per case of 50.6% with 10.5% gain and the best global dice of 72.5% with 11.4% gain.
4 Conclusion
In this work, we have presented a novel importance guided semi-supervised adversarial learning scheme (ISDNet) for medical image segmentation. Specifically, we train two discriminators to enhance the segmentation network with both labeled and unlabeled data to effectively address the insufficient labeled data problem. We combine FCN-based discriminator with CNN-based discriminator for our semi-supervised learning strategy. It can be seen that by integrating these components into our framework, the ISDNet has achieved significant improvement in terms of both accuracy and robustness.
References
Lin, D., Dai, J., Jia, J., et al.: ScribbleSup: scribble-supervised convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3159–3167 (2016)
Brock, A., Donahue, J., Simonyan, K.: Large scale GAN training for high fidelity natural image synthesis. arXiv preprint arXiv:1809.11096 (2018)
Wu, Y., He, K.: Group normalization. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11217, pp. 3–19. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01261-8_1
Tobon-Gomez, C., Geers, A.J., Peters, J., et al.: Benchmark for algorithms segmenting the left atrium from 3D CT and MRI datasets. IEEE Trans. Med. Imaging 34(7), 1460–1473 (2015)
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11211, pp. 833–851. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01234-2_49
Radford, A., Metz, L., Chintala, S.: Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434 (2015)
Nie, D., Gao, Y., Wang, L., Shen, D.: ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 370–378. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_43
Zhang, J., Ding, Z., Li, W., et al.: Importance weighted adversarial nets for partial domain adaptation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8156–8164 (2018)
Frid-Adar, M., Diamant, I., Klang, E., et al.: GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 321, 321–331 (2018)
Hung, W.C., Tsai, Y.H., Liou, Y.T., et al.: Adversarial learning for semi-supervised semantic segmentation. arXiv preprint arXiv:1802.07934 (2018)
Arjovsky, M., Chintala, S., Bottou, L.: Wasserstein GAN. arXiv preprint arXiv:1701.07875 (2017)
Gulrajani, I., Ahmed, F., Arjovsky, M., et al.: Improved training of Wasserstein GANS. In: Advances in Neural Information Processing Systems, pp. 5767–5777 (2017)
Zhu, Y., Elhoseiny, M., Liu, B., et al.: A generative adversarial approach for zero-shot learning from noisy texts. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1004–1013 (2018)
Lai, W.S., Huang, J.B., Ahuja, N., et al.: Deep Laplacian pyramid networks for fast and accurate super-resolution. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 624–632 (2017)
Hu, L., Kan, M., Shan, S., et al.: Duplex generative adversarial network for unsupervised domain adaptation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1498–1507 (2018)
Dai, J., He, K., Sun, J.: BoxSup: exploiting bounding boxes to supervise convolutional networks for semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1635–1643 (2015)
Oh, S.J., Benenson, R., Khoreva, A., et al.: Exploiting saliency for object segmentation from image level labels. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 5038–5047. IEEE (2017)
Pathak, D., Krahenbuhl, P., Darrell, T.: Constrained convolutional neural networks for weakly supervised segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1796–1804 (2015)
Bearman, A., Russakovsky, O., Ferrari, V., Fei-Fei, L.: What’s the point: semantic segmentation with point supervision. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9911, pp. 549–565. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46478-7_34
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Goodfellow, I., Pouget-Abadie, J., Mirza, M., et al.: Generative adversarial nets. In: Advances in Neural Information Processing Systems, pp. 2672–2680 (2014)
Luc, P., Couprie, C., Chintala, S., et al.: Semantic segmentation using adversarial networks. arXiv preprint arXiv:1611.08408 (2016)
Sandoval, Z., Betancur, J., Dillenseger, J.-L.: Multi-atlas-based segmentation of the left atrium and pulmonary veins. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2013. LNCS, vol. 8330, pp. 24–30. Springer, Heidelberg (2014). https://doi.org/10.1007/978-3-642-54268-8_3
Zuluaga, M.A., Cardoso, M.J., Modat, M., Ourselin, S.: Multi-atlas propagation whole heart segmentation from MRI and CTA using a local normalised correlation coefficient criterion. In: Ourselin, S., Rueckert, D., Smith, N. (eds.) FIMH 2013. LNCS, vol. 7945, pp. 174–181. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-38899-6_21
Liu, X., Shen, Y., Zhang, S., et al.: Segmentation of left atrium through combination of deep convolutional and recurrent neural networks. J. Med. Imaging Health Inform. 8(8), 1578–1584 (2018)
Mortazi, A., Karim, R., Rhode, K., Burt, J., Bagci, U.: CardiacNET: segmentation of left atrium and proximal pulmonary veins from MRI using multi-view CNN. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 377–385. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_43
Acknowledgement
This work is supported by: National Natural Science Foundation of China (61673269, 61273285).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ning, Q., Zhao, X., Qian, D. (2019). ISDNet: Importance Guided Semi-supervised Adversarial Learning for Medical Image Segmentation. In: Zhao, Y., Barnes, N., Chen, B., Westermann, R., Kong, X., Lin, C. (eds) Image and Graphics. ICIG 2019. Lecture Notes in Computer Science(), vol 11902. Springer, Cham. https://doi.org/10.1007/978-3-030-34110-7_38
Download citation
DOI: https://doi.org/10.1007/978-3-030-34110-7_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34109-1
Online ISBN: 978-3-030-34110-7
eBook Packages: Computer ScienceComputer Science (R0)