Abstract
3D medical image segmentation with high resolution is an important issue for accurate diagnosis. The main challenge for this task is its large computational cost and GPU memory restriction. Most of the existing 3D medical image segmentation methods are patch-based methods, which ignore the global context information for accurate segmentation and also reduce the efficiency of inference. To tackle this problem, we propose a patch-free 3D medical image segmentation method, which can realize high-resolution (HR) segmentation with low-resolution (LR) input. It contains a multi-task learning framework (Semantic Segmentation and Super-Resolution (SR)) and a Self-Supervised Guidance Module (SGM). SR is used as an auxiliary task for the main segmentation task to restore the HR details, while the SGM, which uses the original HR image patch as a guidance image, is designed to keep the high-frequency information for accurate segmentation. Besides, we also introduce a Task-Fusion Module (TFM) to exploit the inter connections between the segmentation and SR tasks. Since the SR task and TFM are only used in the training phase, they do not introduce extra computational costs when predicting. We conduct the experiments on two different datasets, and the experimental results show that our framework outperforms current patch-based methods as well as has a 4\(\times \) higher speed when predicting. Our codes are available at https://github.com/Dootmaan/PFSeg.
Similar content being viewed by others
Keywords
1 Introduction
Segmentation is one of the most important tasks in medical image analysis. Recent years, with the help of deep learning, there are many inspiring progresses are made in this field. However, most medical images are of high resolution and cannot be directly processed by mainstream graphics cards. Thus, many previous works are 2D networks, which only focus on the segmentation of one single slice at a time [4, 7, 8, 18, 20]. Nevertheless, such methods ignore the valuable information along the z-axis, which limits the improvement of model’s performance. To better capture all the information along the three dimensions, many algorithms such as 2D multiple views [23, 24] and 2.5D [19, 28] are developed, alleviating the problem to some extent. However, these methods still mainly use 2D convolution to extract features and cannot capture the overall 3D spatial information. Therefore, to thoroughly solve this problem, the better way is to use the intuitive 3D convolution [5, 10, 11, 14, 17]. Since training 3D segmentation models needs more computational cost, patch-sampling, that each time only crop a small part from the original medical image as the model’s input, becomes a necessity [15, 21, 25].
Though widely used, patch-sampling also has some flaws. Firstly, the patch-based methods ignore the global context information, which is important for accurate segmentation [27]. As is shown in Fig. 1, our experiments illustrate that the size of a patch can greatly affect the model’s performance, and because bigger patches contain more context, they can usually achieve higher accuracy; Secondly, if the network is trained with patches, it also have to use patches (such as sliding window strategy) in inference stage, which may not only severely decrease the efficiency, but also reduce the accuracy due to inconsistencies introduced during the fusion process that takes place in areas where patches overlap [9].
To solve these problems, we need to design a patch-free segmentation method with moderate computational budgets. Motivated by SR technique, which can recover HR images from LR inputs, we concrete our idea by lowering the resolution of the input image. We propose a novel 3D patch-free segmentation method which can realize HR segmentation with LR input (i.e. the down-sampled 3D image). We call this kind of tasks as Upsample-Segmentation (US) tasks. Inspired by [22], we use SR as an auxiliary task for the US task to restore the HR details lost in the down-sampling procedure. In addition, we introduce a Self-Supervised Guidance Module (SGM), which uses a patch of the original HR image as the subsidiary guidance input. High-frequency features can be directly extracted from it, and then be concatenated with the features from the down-sampled input image. To further improve the model’s performance, we also propose a novel Task-Fusion Module (TFM) to exploit the connections between the US and SR tasks. It should be noted that TFM as well as the auxiliary SR branch are only used in training phase. They do not introduce extra computational costs when predicting.
Our contributions can be mainly concluded into three points: (1) We propose a patch-free 3D medical image segmentation method, which can realize HR segmentation with LR input. (2) We propose a Self-Supervised Guidance Module (SGM), which uses the original HR image patch as guidance, to keep the high-frequency representations for accurate segmentation. (3) We further design a Task-Fusion Module (TFM) to exploit the inter connections between the US and SR tasks, by which the two tasks can be optimized jointly.
2 Methodology
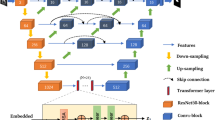
The proposed method is shown in Fig. 2. For a given image, we first down-sample it by 2\(\times \) to lower the resolution, and then use it as the framework’s main input. The encoder will process it into shared features which can be used for both SR and US tasks. In addition, we also crop a patch from the original HR image as guidance, using the features extracted from it to provide the network with more high-frequency information. In training phase, outputs of the US task and SR task will also be sent into Task-Fusion Module (TFM), where the two tasks are fused together to help each other optimize. Note that in the testing phase, only the main segmentation network is used for 3D segmentation, with no extra computational cost.
2.1 Multi-task Learning
Multi-task Learning is the foundation of our framework, including a US task a SR task. Ground truth for US is the labeled segmentation mask of original high resolution, while that of SR is the HR image itself. Since our goal is about generating an accurate segmentation mask, here we will treat US as the main task and SR only as an auxiliary one, which can be removed in testing phase. The two branches are both designed on the basis of ResUNet3D [26] for better consistency, since they share an encoder and will be fused together afterwards. The details are shown in Fig. 3(a).
The loss functions for this part can be divided into segmentation loss \(L_{seg}\) and SR loss \(L_{sr}\) for each task. \(L_{seg}\) consists of Binary Cross Entropy (BCE) Loss and Dice Loss, while \(L_{sr}\) is a simple Mean Square Error (MSE) Loss.
2.2 Self-Supervised Guidance Module
To make proper use of the original HR images and further improve the framework’s performance, we propose the Self-Supervised Guidance Module (SGM). This module uses a typical patch cropped from the original HR image to extract some representative high-frequency features. Through the experiments, we found that simply cropping the central area performs even better than random cropping. Random cropping may cause instability since for every testing case the content of the guidance patch may vary a lot. In our experiment, the size of the guidance patch is set to be 1/64 of the original image.
SGM is designed according to the guidance patch size to make sure the features extracted from it can be correctly concatenated with the shared features. To avoid too much computational cost, SGM is built to be very concise, as is shown in Fig. 3(c). We also introduce a Self-Supervised Guidance Loss (SGL) to evaluate the distance between the guidance and its corresponding part of SR output. The loss function can be described as:
where N refers to the total number of all voxels, and SIG(i) denotes the signal function that will output 1 if i-th voxel is in the cropping window and 0 otherwise. X and \(X\downarrow \) denote the original medical image and the one after down-sampling, while \(SR(\cdot )\) represents the SR output.
2.3 Task-Fusion Module
To better utilize the connections between US and SR, we design a Task-Fusion Module (TFM) combining the two tasks together to let them help each other. This module will first calculate the element-wise product of the two tasks’ outputs (the estimated HR mask and HR image), and then optimize it by two different streams. For the first stream, we propose a Target-Enhanced Loss (TEL), which calculates the average square Euclidean distance of target area voxels. It can be viewed as adding weight to the loss of segmentation target area. Thus, the US task will tend to segment more precisely, and the SR task will pay more attention on the target part. As to the second one, inspired by Spatial Attention Mechanism in [6], we propose a Spatial Similarity Loss (SSL) to make the internal differences between prediction voxels similar to that of the ground truth. SSL is calculated using Spatial Similarity Matrix, which mainly describes the pairwise relationship between voxels. For a \(D\times W\times H\times C\) image I (for medical images C usually equals 1), to compute its Spatial Similarity Matrix, first we need to reshape it into \(V\times C\), where \(V=D\times W\times H\). After that, by multiplying this matrix with its transpose, we can have the \(V\times V\) similarity matrix and calculate the loss of it with ground truth. The loss function for this module can be defined as follows.
where \(p_i\) denotes the prediction of i-th voxel after binarization, and \(y_i\) represents its corresponding ground truth. \(S_{ij}\) refers to the correlation between i-th and j-th voxel of fusion image I, while \(I_i\) represents the i-th voxel of the image.
2.4 Overall Objective Function
The overall objective function L of the proposed framework is:
where \(\omega _{sr}\), \(\omega _{tfm}\) and \(\omega _{sgl}\) are hyper-parameters, and are all set to 0.5 by default. The whole objective function can be optimized end-to-end.
3 Experiments
3.1 Datasets
We used BRATS2020 dataset [2, 3, 16] and a privately-owned liver segmentation dataset in the experiment. BRATS2020 dataset contains a total number of 369 subjects, each with four-modality MRI images (T1, T2, T1ce and FLAIR) of size \(240\times 240\times 155\) and spacing \(1\times 1\times 1\) mm\(^3\). The ground truth includes masks of Tumor Core (TC), Enhanced Tumor (ET) and Whole Tumor (WT). For each image, we removed the edges without brain part by 24 voxels and resized the rest part to resolution \(192\times 192\times 128\). In our experiment, we used the down- sampled T2-weighted images as input, the original T2-weighted images as SR ground truth, and WT masks as US ground truth.
The privately-owned liver segmentation dataset contains 347 subjects. Each one has an MRI image and a segmentation ground truth labeled by experienced doctors. In our experiment, spacing of the images were all regulated to \(1.5\times 1.5\times 1.5\) mm\(^3\), and we then cropped the central \(192\times 192\times 128\) area. The cropped MRI image and its segmentation mask are used as ground truth of SR and US, while the input is the cropped image after down-sampling.
3.2 Implementation Details
We compared our framework with different patch-based 3D segmentation models. For those methods, we predict the test image using sliding window strategy with a stride of 48, 48 and 32 for x-axis, y-axis and z-axis, respectively. Besides, we also tested our method with other patch-free segmentation models (i.e., ResUNet3D\(\uparrow \) and HDResUNet). ResUNet3D\(\uparrow \) conducts ordinary segmentation with a down-sampled image, then enlarging the result by tricubic interpolation [13]; HDResUNet uses Holistic Decomposition [27] with ResUNet3D, and the down-shuffling factors of it are all set to 2.
We employed three quantitative evaluation indices in the experiments, which are Dice Similarity Coefficient, 95% Hausdorff Distance and Jaccard Coefficient. Dice and Jaccard mainly focus on the segmentation area. 95% Hausdorff pays more attention to the edges.
All the experiments run on a Nvidia GTX 1080Ti GPU with 11 GB video memory. For fair comparison, the input sizes were all set to \(96\times 96\times 64\), except HDResUNet, which uses the original HR image. Therefore, the patch size for patch-based methods and the input image size for patch-free methods are the same. For both datasets, we used 80% for training and the rest for testing. Data augmentation includes random cropping (only for patch-based methods), random flip, random rotation, and random shift. All the models are optimized by Adam [12] with the initial learning rate set to \(1e^{-4}\). The rate will be divided by 10 if the loss does not continuously reduce over 20 epochs, and the training phase will end when it reaches \(1e^{-7}\).
3.3 Ablation Study
We conduct an ablation study on BRATS2020 to investigate how the designed modules affect the framework’s performance. The framework is tested with two different backbones (i.e., UNet3D and ResUNet3D).
In Table 1, for both backbones, appending SR as the auxiliary task improves the segmentation performance, indicating that the framework successfully rebuild some high-frequency information with the help of SR. Moreover, the segmentation result after introducing TEL and SSL also proves the effectivity of TFM, showing that the inter connection between US and SR is very useful for joint optimization. At last, the increase of all the metrics after adding SGM demonstrate that the framework benefits from the self-supervised guidance for the high-frequency features it brings.
3.4 Experimental Results
The experimental results are summarized in Table 2. Our framework surpasses traditional 3D patch-based methods and also outperforms the other patch-free methods. Patch-free methods have the most obvious improvements in 95% Hausdorff Distance: with the global context, the model can more easily segment the target area as a whole, hence making the segmentation edges smoother and more accurate. Since our framework can directly output a complete segmentation mask at a time, it also has a faster inference speed than most of the other methods.
Some typical segmentation results are listed in Fig. 4. As is shown, the patch-based results have many obvious flaws (labeled in red): in Case1, there is some segmentation noise. This problem mainly results from the limited context in patches. When conducting segmentation on the upper right corner patch, the model does not have the information of the real tumor area and it will be more likely to misdiagnose normal area as lesion. In Case2 and Case3, there are some failed segmentation in the corner area due to the padding technique. In [1], the authors pointed out that padding may result in artifacts on the edges of feature maps, and these artifacts may confuse the network. The problem of Case2 and Case3 is commonly seen when target area leaps over several patches. Under such circumstances, it is difficult for the patch-based models to correctly estimate the voxels on the edges, hence resulting in inconsistencies during the fusion process. Although patch-free segmentation can solve the above-mentioned problems, it may lead to significant performance degradation due to the loss of high-frequency information during down-scaling. In our method, we build a multi-task learning framework (US and SR) with two well-designed modules (TFM and SGM) to keep the HR representations, thus avoiding this issue. Therefore, our framework outperforms other existing patch-free methods.
4 Conclusion
In this work, we propose a novel framework for fast and accurate patch-free segmentation, which is capable of capturing global context while not introducing too much extra computational cost. We validate the framework’s performance on two datasets to demonstrate its effectiveness, and the result shows that it can efficiently generate better segmentation mask than other patch-based and patch-free methods.
References
Alsallakh, B., Kokhlikyan, N., Miglani, V., Yuan, J., Reblitz-Richardson, O.: Mind the pad - cnns can develop blind spots. In: International Conference on Learning Representations (2021). https://openreview.net/forum?id=m1CD7tPubNy
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J.S., Freymann, J.B., Farahani, K., Davatzikos, C.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. data 4(1), 1–13 (2017)
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint arXiv:1811.02629 (2018)
Christ, P.F., et al.: Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 415–423. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_48
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Fu, J., et al.: Dual attention network for scene segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 3146–3154 (2019)
Huang, H., et al.: Unet 3+: a full-scale connected unet for medical image segmentation. In: ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059. IEEE (2020)
Huang, H., et al.: Medical image segmentation with deep atlas prior. IEEE Trans. Med. Imaging (2021)
Huang, Y., Shao, L., Frangi, A.F.: Simultaneous super-resolution and cross-modality synthesis of 3D medical images using weakly-supervised joint convolutional sparse coding. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6070–6079 (2017)
Kao, P.Y., et al.: Improving patch-based convolutional neural networks for MRI brain tumor segmentation by leveraging location information. Front. Neurosci. 13, 1449 (2020)
Kim, H., et al.: Abdominal multi-organ auto-segmentation using 3d-patch-based deep convolutional neural network. Sci. Rep. 10(1), 1–9 (2020)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: International Conference on Learning Representations (2015)
Lekien, F., Marsden, J.: Tricubic interpolation in three dimensions. Int. J. Numer. Methods Eng. 63(3), 455–471 (2005)
Li, Z., Pan, J., Wu, H., Wen, Z., Qin, J.: Memory-efficient automatic kidney and tumor segmentation based on non-local context guided 3D U-Net. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12264, pp. 197–206. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59719-1_20
Madesta, F., Schmitz, R., Rösch, T., Werner, R.: Widening the focus: biomedical image segmentation challenges and the underestimated role of patch sampling and inference strategies. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12264, pp. 289–298. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59719-1_29
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Milletari, F., Navab, N., Ahmadi, S.A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Shao, Q., Gong, L., Ma, K., Liu, H., Zheng, Y.: Attentive CT lesion detection using deep pyramid inference with multi-scale booster. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 301–309. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_34
Tang, Y., Tang, Y., Zhu, Y., Xiao, J., Summers, R.M.: E\(^2\)Net: an edge enhanced network for accurate liver and tumor segmentation on CT scans. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12264, pp. 512–522. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59719-1_50
Tang, Y., et al.: High-resolution 3D abdominal segmentation with random patch network fusion. Med. Image Anal. 69, 101894 (2021)
Wang, L., Li, D., Zhu, Y., Tian, L., Shan, Y.: Dual super-resolution learning for semantic segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 3774–3783 (2020)
Wang, Y., Zhou, Y., Shen, W., Park, S., Fishman, E.K., Yuille, A.L.: Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Med. Image Anal. 55, 88–102 (2019)
Xia, Y., et al.: Uncertainty-aware multi-view co-training for semi-supervised medical image segmentation and domain adaptation. Med. Image Anal. 65, 101766 (2020)
Yang, H., Shan, C., Bouwman, A., Kolen, A.F., de With, P.H.: Efficient and robust instrument segmentation in 3D ultrasound using patch-of-interest-fusenet with hybrid loss. Med. Image Anal. 67, 101842 (2021)
Yu, L., Yang, X., Chen, H., Qin, J., Heng, P.A.: Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 31 (2017)
Zeng, G., Zheng, G.: Holistic decomposition convolution for effective semantic segmentation of medical volume images. Med. Image Anal. 57, 149–164 (2019)
Zlocha, M., Dou, Q., Glocker, B.: Improving RetinaNet for CT lesion detection with dense masks from weak RECIST labels. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 402–410. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_45
Acknowledgement
This work was supported in part by Major Scientific Research Project of Zhejiang Lab under the Grant No. 2020ND8AD01, and in part by the Grant-in Aid for Scientific Research from the Japanese Ministry for Education, Science, Culture and Sports (MEXT) under the Grant No. 20KK0234, No. 21H03470, and No. 20K21821.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, H. et al. (2021). Patch-Free 3D Medical Image Segmentation Driven by Super-Resolution Technique and Self-Supervised Guidance. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12901. Springer, Cham. https://doi.org/10.1007/978-3-030-87193-2_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-87193-2_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87192-5
Online ISBN: 978-3-030-87193-2
eBook Packages: Computer ScienceComputer Science (R0)