Abstract
Aim: The development and evaluation of deep learning (DL) and radiomics based models for recurrence-free survival (RFS) prediction in oropharyngeal squamous cell carcinoma (OPSCC) patients based on clinical features, positron emission tomography (PET) and computed tomography (CT) scans and GTV (Gross Tumor Volume) contours of primary tumors and pathological lymph nodes.
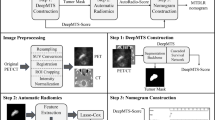
Methods: A DL auto-segmentation algorithm generated the GTV contours (task 1) that were used for imaging biomarkers (IBMs) extraction and as input for the DL model. Multivariable cox regression analysis was used to develop radiomics models based on clinical and IBMs features. Clinical features with a significant correlation with the endpoint in a univariable analysis were selected. The most promising IBMs were selected by forward selection in 1000 times bootstrap resampling in five-fold cross validation. To optimize the DL models, different combinations of clinical features, PET/CT imaging, GTV contours, the selected radiomics features and the radiomics model predictions were used as input. The combination with the best average performance in five-fold cross validation was taken as the final input for the DL model. The final prediction in the test set, was an ensemble average of the predictions from the five models for the different folds.
Results: The average C-index in the five-fold cross validation of the radiomics model and the DL model were 0.7069 and 0.7575, respectively. The radiomics and final DL models showed C-indexes of 0.6683 and 0.6455, respectively in the test set.
Conclusion: The radiomics model for recurrence free survival prediction based on clinical, GTV and CT image features showed the best predictive performance in the test set with a C-index of 0.6683.
B. Ma and Y. Li—These authors contributed equally.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
World Health Organization: Global cancer observatory. International agency for research on cancer. World Health Organization (2020)
O’Sullivan, B., et al.: Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol. 17(4) (2016)
Cramer, J.D., Burtness, B., Le, Q.T., Ferris, R.L.: The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 16(11) (2019)
Ma, B., et al.: Self-supervised multi-modality image feature extraction for the progression free survival prediction in head and neck cancer. In: Andrearczyk, V., Oreiller, V., Hatt, M., Depeursinge, A. (eds.) HECKTOR 2021. LNCS, vol. 13209, pp. 308–317. Springer, Cham (2022). https://doi.org/10.1007/978-3-030-98253-9_29
Zhai, T.T., et al.: The prognostic value of CT-based image-biomarkers for head and neck cancer patients treated with definitive (chemo-)radiation. Oral Oncol. 95 (2019)
Zhai, T.T., et al.: Improving the prediction of overall survival for head and neck cancer patients using image biomarkers in combination with clinical parameters. Radiother. Oncol. 124(2) (2017)
Bi, W.L., et al.: Artificial intelligence in cancer imaging: clinical challenges and applications. CA: Cancer J. Clin. (2019)
Ma, B., et al.: MRI image synthesis with dual discriminator adversarial learning and difficulty-aware attention mechanism for hippocampal subfields segmentation. Comput. Med. Imaging Graph. 86 (2020)
Zhao, Y., Ma, B., Jiang, P., Zeng, D., Wang, X., Li, S.: Prediction of Alzheimer’s disease progression with multi-information generative adversarial network. IEEE J. Biomed. Heal. Inform. 25(3) (2021)
Zhang, X., Kelkar, V.A., Granstedt, J., Li, H., Anastasio, M.A.: Impact of deep learning-based image super-resolution on binary signal detection. J. Med. Imaging 8(06) (2021)
Zeng, D., Li, Q., Ma, B., Li,S.: Hippocampus segmentation for preterm and aging brains using 3D densely connected fully convolutional networks. IEEE Access 8 (2020)
Oreiller, V., et al.: Head and neck tumor segmentation in PET/CT: the HECKTOR challenge. Med. Image Anal. 77, 102336 (2022)
Diamant, A., Chatterjee, A., Vallières, M., Shenouda, G., Seuntjens, J.: Deep learning in head & neck cancer outcome prediction. Sci. Rep. 9(1) (2019)
Wang, Y., et al.: Deep learning based time-to-event analysis with PET, CT and joint PET/CT for head and neck cancer prognosis. Comput. Methods Programs Biomed. 106948 (2022)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, vol. 2016-December (2016)
Andrearczyk, V., et al.: Overview of the HECKTOR challenge at MICCAI 2021: automatic head and neck tumor segmentation and outcome prediction in PET/CT images. In: Andrearczyk, V., Oreiller, V., Hatt, M., Depeursinge, A. (eds.) HECKTOR 2021. LNCS, vol. 13209, pp. 1–37. Springer, Cham (2022). https://doi.org/10.1007/978-3-030-98253-9_1
Van Griethuysen, J.J.M., et al.: Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77(21) (2017)
van Dijk, L.V., et al.: 18F-FDG PET image biomarkers improve prediction of late radiation-induced xerostomia. Radiother. Oncol. 126(1), 89–95 (2018)
van Dijk, L.V., et al.: CT image biomarkers to improve patient-specific prediction of radiation-induced xerostomia and sticky saliva. Radiother. Oncol. 122(2), 185–191 (2017)
Van den Bosch, L., et al.: Key challenges in normal tissue complication probability model development and validation: towards a comprehensive strategy. Radiother. Oncol. 148 (2020)
Katzman, J.L., Shaham, U., Cloninger, A., Bates, J., Jiang, T., Kluger, Y.: DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18(1) (2018)
Chamberlain, C., Owen-Smith, A., Donovan, J., Hollingworth, W.: A systematic review of geographical variation in access to chemotherapy. BMC Cancer 16(1) (2015)
Leijenaar, R.T.H., et al.: Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br. J. Radiol. 91(1086), 1–8 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Ma, B. et al. (2023). Deep Learning and Radiomics Based PET/CT Image Feature Extraction from Auto Segmented Tumor Volumes for Recurrence-Free Survival Prediction in Oropharyngeal Cancer Patients. In: Andrearczyk, V., Oreiller, V., Hatt, M., Depeursinge, A. (eds) Head and Neck Tumor Segmentation and Outcome Prediction. HECKTOR 2022. Lecture Notes in Computer Science, vol 13626. Springer, Cham. https://doi.org/10.1007/978-3-031-27420-6_24
Download citation
DOI: https://doi.org/10.1007/978-3-031-27420-6_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-27419-0
Online ISBN: 978-3-031-27420-6
eBook Packages: Computer ScienceComputer Science (R0)