Abstract
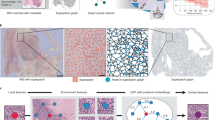
Many recent methods for the analysis of histology whole slide images (WSIs) have used graph neural networks (GNNs) to aggregate visual information over a large image resolution. However, domain shift is a significant challenge in computational histopathology, due to differences in WSI appearance between institutes, and the effect of these differences on training GNNs has not been explored. In this work, we present the Multiple Embedding Graph Augmentation (MEGA) strategy to improve the cross-institute generalisation of GNNs in histology. We show that by introducing image augmentation and normalisation to the node features used to train a GNN, we can train a model that is robust to domain shift without additional labels or further training of the feature extractor. We compare MEGA to noise-based regularisation and demonstrate its effectiveness in a node classification tissue prediction task in placenta histology.
J. Campbell and C. Vanea—Contributed equally to this work.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ahmedt-Aristizabal, D., Armin, M.A., Denman, S., Fookes, C., Petersson, L.: A survey on graph-based deep learning for computational histopathology. Comput. Med. Imaging Graph. 95, 102027 (2022). https://doi.org/10.1016/j.compmedimag.2021.102027
Bándi, P., et al.: From detection of individual metastases to classification of lymph node status at the patient level: the CAMELYON17 challenge. IEEE Trans. Med. Imaging 38(2), 550–560 (2019). Conference Name: IEEE Transactions on Medical Imaging. https://doi.org/10.1109/TMI.2018.2867350
Cai, T., Luo, S., Xu, K., He, D., Liu, T.Y., Wang, L.: GraphNorm: a principled approach to accelerating graph neural network training. In: Meila, M., Zhang, T. (eds.) Proceedings of the 38th International Conference on Machine Learning. Proceedings of Machine Learning Research, July 2021, vol. 139, pp. 1204–1215. PMLR (2021)
Chiang, W.L., Liu, X., Si, S., Li, Y., Bengio, S., Hsieh, C.J.: Cluster-GCN: an efficient algorithm for training deep and large graph convolutional networks. In: Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, KDD 2019, pp. 257–266. Association for Computing Machinery, New York (2019). Event-place: Anchorage, AK, USA. https://doi.org/10.1145/3292500.3330925
Chlipala, E.A., et al.: Impact of preanalytical factors during histology processing on section suitability for digital image analysis. Toxicol. Pathol. 49(4), 755–772 (2021). Publisher: SAGE Publications Inc. https://doi.org/10.1177/0192623320970534
Cubuk, E.D., Zoph, B., Shlens, J., Le, Q.V.: RandAugment: practical automated data augmentation with a reduced search space. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, June 2020, pp. 3008–3017. IEEE (2020). https://doi.org/10.1109/CVPRW50498.2020.00359
Ding, K., Xu, Z., Tong, H., Liu, H.: Data augmentation for deep graph learning: a survey. SIGKDD Explor. Newsl. 24(2), 61–77 (2022). Place: New York, NY, USA Publisher: Association for Computing Machinery. https://doi.org/10.1145/3575637.3575646
Faryna, K., van der Laak, J., Litjens, G.: Automatic data augmentation to improve generalization of deep learning in H &E stained histopathology. Comput. Biol. Med. 170, 108018 (2024). https://doi.org/10.1016/j.compbiomed.2024.108018
Faryna, K., Laak, J., Litjens, G.: Tailoring automated data augmentation to H &E-stained histopathology. In: Proceedings of the Fourth Conference on Medical Imaging with Deep Learning, August 2021, pp. 168–178. PMLR (2021). ISSN 2640-3498
Godwin, J., et al.: Simple GNN regularisation for 3D molecular property prediction & beyond. arXiv arXiv:2106.07971 [cs], March 2022.https://doi.org/10.48550/arXiv.2106.07971
Hoffman, J., et al.: CyCADA: cycle-consistent adversarial domain adaptation. In: Proceedings of the 35th International Conference on Machine Learning, July 2018, pp. 1989–1998. PMLR (2018). ISSN 2640-3498
Kang, H., et al.: StainNet: a fast and robust stain normalization network. Front. Med. 8, 746307 (2021)
Kong, K., et al.: Robust optimization as data augmentation for large-scale graphs. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), June 2022. pp. 60–69 (2022)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, vol. 25. Curran Associates, Inc. (2012)
Lim, S., Kim, I., Kim, T., Kim, C., Kim, S.: Fast autoaugment. In: Advances in Neural Information Processing Systems, vol. 32. Curran Associates, Inc. (2019)
McInnes, L., Healy, J., Saul, N., Großberger, L.: UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3(29), 861 (2018). https://doi.org/10.21105/joss.00861
Papp, P.A., Martinkus, K., Faber, L., Wattenhofer, R.: DropGNN: random dropouts increase the expressiveness of graph neural networks. arXiv arXiv:2111.06283 [cs], November 2021. https://doi.org/10.48550/arXiv.2111.06283
Rong, Y., Huang, W., Xu, T., Huang, J.: DropEdge: towards deep graph convolutional networks on node classification. arXiv arXiv:1907.10903 [cs, stat], March 2020. https://doi.org/10.48550/arXiv.1907.10903,
Ruifrok, A.C., Johnston, D.A.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23(4), 291–299 (2001)
Runz, M., Rusche, D., Schmidt, S., Weihrauch, M.R., Hesser, J., Weis, C.A.: Normalization of HE-stained histological images using cycle consistent generative adversarial networks. Diagn. Pathol. 16(1), 71 (2021). https://doi.org/10.1186/s13000-021-01126-y
Shaban, M.T., Baur, C., Navab, N., Albarqouni, S.: StainGAN: stain style transfer for digital histological images. arXiv arXiv:1804.01601 [cs], April 2018. https://doi.org/10.48550/arXiv.1804.01601
Shorten, C., Khoshgoftaar, T.M.: A survey on image data augmentation for deep learning. J. Big Data 6(1), 60 (2019). https://doi.org/10.1186/s40537-019-0197-0
Stacke, K., Eilertsen, G., Unger, J., Lundström, C.: Measuring domain shift for deep learning in histopathology. IEEE J. Biomed. Health Inf. 25(2), 325–336 (2021). Conference Name: IEEE Journal of Biomedical and Health Informatics. https://doi.org/10.1109/JBHI.2020.3032060
Studer, L., Wallau, J., Dawson, H., Zlobec, I., Fischer, A.: Classification of intestinal gland cell-graphs using graph neural networks. In: 2020 25th International Conference on Pattern Recognition (ICPR), pp. 3636–3643, January 2021. ISSN 1051-4651. https://doi.org/10.1109/ICPR48806.2021.9412535
Tellez, D., et al.: Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 58, 101544 (2019). https://doi.org/10.1016/j.media.2019.101544
Theissen, H., Chakraborty, T., Malacrino, S., Royston, D., Rittscher, J.: Multi-scale graphical representation of cell environment. In: 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), July 2022, pp. 3522–3525 (2022). ISSN 2694-0604. https://doi.org/10.1109/EMBC48229.2022.9871710
Vanea, C., et al.: Mapping cell-to-tissue graphs across human placenta histology whole slide images using deep learning with HAPPY. Nat. Commun. 15(1), 2710 (2024). https://doi.org/10.1038/s41467-024-46986-2
Wang, J., Chen, R.J., Lu, M.Y., Baras, A., Mahmood, F.: Weakly supervised prostate TMA classification via graph convolutional networks. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), April 2020, pp. 239–243 (2020). ISSN 1945-8452. https://doi.org/10.1109/ISBI45749.2020.9098534
Wang, Y., Wang, W., Liang, Y., Cai, Y., Liu, J., Hooi, B.: NodeAug: semi-supervised node classification with data augmentation. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Event, CA, USA, August 2020, pp. 207–217. ACM (2020). https://doi.org/10.1145/3394486.3403063
Wang, Z., et al.: Cross-domain nuclei detection in histopathology images using graph-based nuclei feature alignment. IEEE J. Biomed. Health Inf. 28(1), 78–88 (2024). Conference Name: IEEE Journal of Biomedical and Health Informatics. https://doi.org/10.1109/JBHI.2023.3280958
Wu, Q., Zhang, H., Yan, J., Wipf, D.: Handling distribution shifts on graphs: an invariance perspective. arXiv arXiv:2202.02466 [cs], May 2022. https://doi.org/10.48550/arXiv.2202.02466
Yang, J., Chen, H., Yan, J., Chen, X., Yao, J.: Towards better understanding and better generalization of few-shot classification in histology images with contrastive learning. arXiv arXiv:2202.09059 [cs, eess], February 2022. https://doi.org/10.48550/arXiv.2202.09059
Zhao, T., et al.: Graph data augmentation for graph machine learning: a survey. arXiv arXiv:2202.08871 [cs], January 2023. https://doi.org/10.48550/arXiv.2202.08871
Zhao, Y., et al.: Predicting lymph node metastasis using histopathological images based on multiple instance learning with deep graph convolution. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), June 2020, pp. 4836–4845 (2020). ISSN 2575-7075. https://doi.org/10.1109/CVPR42600.2020.00489
Zheng, Y., et al.: A graph-transformer for whole slide image classification. IEEE Trans. Med. Imaging 41(11), 3003–3015 (2022). https://doi.org/10.1109/TMI.2022.3176598
Zhou, Y., Graham, S., Koohbanani, N.A., Shaban, M., Heng, P.A., Rajpoot, N.: CGC-Net: cell graph convolutional network for grading of colorectal cancer histology images. arXiv arXiv:1909.01068 [cs, eess], September 2019.https://doi.org/10.48550/arXiv.1909.01068
Zhu, Q., Ponomareva, N., Han, J., Perozzi, B.: Shift-Robust GNNs: overcoming the limitations of localized graph training data. arXiv arXiv:2108.01099 [cs], October 2021. https://doi.org/10.48550/arXiv.2108.01099
Acknowledgments
Jonathan Campbell and Claudia Vanea are supported by the EPSRC Center for Doctoral Training in Health Data Science (EP/S02428X/1).
Cecilia M. Lindgren is supported by the Li Ka Shing Foundation, NIHR Oxford Biomedical Research Centre, Oxford, NIH (1P50HD104224-01), Gates Foundation (INV-024200), and a Wellcome Trust Investigator Award (221782/Z/20/Z).
Triin Lasik is funded by the European Regional Development Fund, the programme Mobilitas Pluss (MOBTP155) and the Estonian Research Council grant (PSG776).
The computational aspects of this research were supported by the Wellcome Trust Core Award (203141/Z/16/Z) and the NIHR Oxford BRC. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
We thank Andrew Zisserman for his comments and suggestions.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors have no competing interests to declare that are relevant to the content of this article.
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
A Training Data Augmentation for Cell Model
In Table 4, we list the augmentations used to train the cell classifier used as the feature extractor.
B Whole Slide Image Dataset Details
Slides were prepared using a standard formalin fixing, paraffin-embedded, and hematoxylin and eosin staining procedure, sliced at 5 \(\upmu \)m full thickness, and scanned at \(\times \)40 magnification.
Source Institute. The source slides used for training the feature extractor and GNN models were collected at the University of Tartu, Estonia, and digitised using a Hamamatsu XR scanner.
Target Institute 1. Two target slides used for evaluating the generalisability of the GNN model were collected at the Northshore University HealthSystem, Chicago, and digitised using an Aperio GT 450 scanner.
Target Institute 2. Two target slides used for evaluating the generalisability of the GNN model were collected at the Hadassah Medical Center, Israel, and digitised using a 3D HISTECH PANNORAMIC 250 Flash III.
C Node Classification Dataset Details
Source Institute. 789,539 nodes from the source WSI were manually labelled into 9 tissue classes with a 55%/24%/21% train/val/test split to train and evaluate the performance of the GNN for node classification. Dataset splits were chosen as large, consistent regions with similar class distributions to avoid information leakage between neighbouring nodes.
Target Institute 1. 39,094 nodes from two target WSIs were manually labelled into 9 tissue classes to evaluate the generalisation of the GNN for node classification. Evaluation nodes were selected from different regions of the slide and have a similar class distribution to the training data.
Target Institute 2. 65,892 nodes from two target WSIs were manually labelled into 9 tissue classes to evaluate the generalisation of the GNN for node classification. Evaluation nodes were selected from different regions of the slide and have a similar class distribution to the training data.
D Feature Extractor Dataset Details
Source Institute. 11,955 \(200\times 200\) pixel images of 11 cell types from five slides, separate from those used for the GNN, were manually labelled into a 72%/14%/14% train/val/test split to train and evaluate the performance of the feature extractor on the source data.
Target Institute 1. 361 \(200\times 200\) pixel images of 11 cell types from two slides, separate from those used for the GNN, were manually labelled to quantify the degree of domain shift by the feature extractor performance on the target 1 data. For training the unbiased feature extractor, 2,415 \(200\times 200\) pixel images of cells from 2 slides were manually labelled into a 70%/15%/15% train/val/test split.
Target Institute 2. 686 \(200\times 200\) pixel images of 11 cell types from two slides, separate from those used for the GNN, were manually labelled to quantify the degree of domain shift by the feature extractor performance on the target 2 data.
E Training Details
For the cell classifier feature extractor, we finetune a ResNet-50 model with additional linear layers from ImageNet weights for 60 epochs with an Adam optimiser, 400 batch size, cross entropy-loss, and a 0.0001 learning rate with 0.5 weight decay applied every 20 epochs. Minority classes are oversampled during training. The model with the highest validation accuracy is then fully trained unfrozen for 100 epochs with the same hyperparameters. The model is trained to predict the cell class of the cell in the centre of a \(200\times 200\) pixel image. Cells are classified into one of 11 cell types: syncytiotrophoblast, cytotrophoblast, syncytial knot, extravillous trophoblast, undifferentiated mesenchymal cell, fibroblast, vascular endothelial cell, vascular myocyte, Hofbauer cell, maternal decidual cell and leukocytes.
For the GNN, we train a randomly initialised ClusterGCN model with 16 GraphSAGEConv layers, each with 256 hidden units for 2000 epochs with an Adam optimiser, cross entropy-loss, a 0.001 learning rate, a batch size of 200 and a subgraph sampling size of 400 neighbours. The model with the highest validation accuracy, calculated without neighbourhood sampling, is saved as the final model. The GNN is trained to classify each node into one of 9 tissue types: stem villi, anchoring villi, mature intermediate villi, terminal villi, villus sprouts, chorionic plate, basal plate, fibrin and avascular villi.
F Results on Additional Target Slides
We randomly select and annotate nodes in an additional target slide from each target institute to further assess the improvements in generalisation from each method (Table 5). Node annotations are made such that they maintain a similar class distribution to the evaluation sets from other slides. Using the naive results as a measure of domain shift, we see that when there isn’t a domain shift, such as between source and target 2, then graph normalisation will provide the most benefit. When there is a domain shift, such as between source and target 1, then norm and pixel augmentations achieve the best performance. These results are consistent with the reported accuracies on the other target slides and when using an unbiased feature extractor.
G Hardware and Software Details
All training and inference were performed on a single NVIDIA A100 GPU. Cell images were extracted from WSIs using libvips v8.9.2 with OpenSlide v3.4.1 via Python bindings pyvips v2.1.14. The code was written in Python v3.10.13 with PyTorch v2.0.1. torchvision v0.15.2, and PyTorch Geometric v2.3.1 for the deep learning models. Augmentations were applied using Albumentations v1.3.0. WSI cell and tissue annotations were created using QuPath v0.3.1.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Campbell, J. et al. (2024). Enhancing Cross-Institute Generalisation of GNNs in Histopathology Through Multiple Embedding Graph Augmentation (MEGA). In: Yap, M.H., Kendrick, C., Behera, A., Cootes, T., Zwiggelaar, R. (eds) Medical Image Understanding and Analysis. MIUA 2024. Lecture Notes in Computer Science, vol 14860. Springer, Cham. https://doi.org/10.1007/978-3-031-66958-3_20
Download citation
DOI: https://doi.org/10.1007/978-3-031-66958-3_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-66957-6
Online ISBN: 978-3-031-66958-3
eBook Packages: Computer ScienceComputer Science (R0)