Abstract
Three-dimensional reconstructions of coronary arteries can overcome some of the limitations of 2D X-ray angiography, namely artery overlap/foreshortening and lack of depth information. Model-based arterial reconstruction algorithms usually rely on 2D coronary artery segmentations and require good robustness to outliers. In this paper, we propose a novel probabilistic method to reconstruct coronary artery centrelines from retrospectively gated X-ray images based on a probabilistic mixture model. Specifically, 3D coronary artery centrelines are described by a mixture of Student’s t-distributions, and the reconstruction is formulated as maximum-likelihood estimation of the mixture model parameters, given the 2D segmentations of arteries from 2D X-ray images. Our method provides robustness against the erroneously segmented parts in the 2D segmentations by taking advantage of the inherent robustness of t-distributions. We validate our reconstruction results using synthetic phantom and clinical X-ray angiography data. The results show that the proposed method can cope with imperfect and noisy segmentation data.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
X-ray coronary angiography is one of the commonly utilized imaging modalities in the assessment of coronary artery disease. However, this modality is known to be limited, since it can only deliver 2D X-ray images to visualise 3D moving coronary arteries. To overcome this limitation, 3D description of the coronary arteries can be reconstructed from X-ray angiography images [1]. However, inverse problem of reconstruction remains a challenging task due to intensity inhomogeneities, artery overlap/foreshortening, and cardiac/respiratory motion.
Numerous methods have been proposed to reconstruct coronary artery trees from X-ray angiography. Among these methods, model-based reconstruction methods try to reconstruct a 3D representation of the coronary arteries that comprises of artery centrelines and, occasionally, the arterial lumen surface. Most of the existing model-based reconstruction methods require clean segmentations of arteries from 2D X-ray angiography images [1]. However, segmentation of coronary arteries from X-ray angiography is still a challenging task because of inhomogeneous intensities and artery overlap/foreshortening, thus prone to errors. Therefore, the reconstruction methods should be devised in a way that they become robust to possible errors in the 2D coronary artery segmentations (such as over-segmentations due to the other structures that are also visible in the X-ray angiography images).
In this paper, we propose a novel method to reconstruct coronary artery centrelines from retrospectively gated X-ray angiography images acquired via a calibrated X-ray angiography system. Our method employs a novel probabilistic formulation based on a mixture of Student’s t-distributions that describes the coronary artery centrelines in 3D space. Given the 2D over-segmentations of arteries from 2D X-ray images, we formulate the reconstruction problem as maximum-likelihood (ML) estimation of mixture model parameters. The t-distributions are known to be inherently robust to the outliers in the data. The utilization of t-distributions as the components of the mixture model allows us to significantly reduce the burden of manual inspecting the segmented arteries. This opens the possibility to use the results from semi/automatic 2D vascular segmentation algorithms, facilitating the reconstruction process. To the best of our knowledge, this is the first paper proposing that the reconstruction problem can be alternatively viewed as the task of a Student’s t-mixture model fitting. Apart from robustness, the proposed method is a versatile framework that can handle complex arterial geometries, and requires no point-to-point correspondences.
2 Method
Our method assumes that the X-ray images and a simultaneous ECG are collected using a calibrated angiography system. Coronary artery segmentations, possibly containing erroneously segmented parts, are extracted from retrospectively gated 2D X-ray images. Coronary arteries are represented by a mixture of Student’s t-distributions in 3D, from which the points in the 2D segmentations are considered to be generated. Reconstruction is formulated as a ML estimation of the parameters of the mixture model.
X-ray Image Acquisition and Retrospective ECG Gating: Our method requires an X-ray acquisition by a calibrated angiography system, i.e. the system geometry is known for each X-ray projection. This information can be exploited to define the projection between the patient and X-ray detector coordinate systems, which can be modelled by weak-perspective or perspective camera models [3]. Weak-perspective camera model provides a linear approximation of perspective camera model, when the depth of the object is small compared to distance from camera and the field of view is small [3], which are valid for X-ray angiography. Linearity of weak-perspective approximation yields closed form solutions for the estimation of mixture model parameters. Our method benefits from both camera models, starting with a weak-perspective model and, upon convergence, switching to perspective model for refinement.
Respiratory motion can be reduced by collecting X-ray images during a breath hold. Following image acquisition, a subset of X-ray images are selected via retrospective ECG gating in order to compensate for the cardiac motion. We select frames that are closest to a cardiac phase with minimal motion.
Segmentation of X-ray Images: The coronary arteries are segmented from 2D X-ray images using an automatic segmentation algorithm such as the ones proposed in [5, 8]. The resulting segmentation may include some parts, which are erroneously segmented due to noise or other structures such as catheter, spine and diaphragm. We refer to these erroneous parts of the segmentation as outliers. Next, the segmented arteries are converted to point sets and further processed by our algorithm.
Reconstruction Based on Mixture of Student’s t-distributions: To formulate the reconstruction, we represent 3D coronary artery centrelines by a probabilistic mixture model. Specifically, the 3D centrelines can be described by a set of points in 3D space. Furthermore, spatial locations of these points can be defined by the mean values of mixture model components. As a result, the points describing the 3D coronary artery centrelines form a mixture distribution in 3D space. We opt for Student’s t-distribution as the component distribution of the mixture model, which is known to be a robust alternative to Gaussian distribution in the presence of outlier samples, owing to its heavier tails [6].
Segmented 2D artery points on X-ray images are considered to be the projections of samples generated from mixture model distribution. Therefore, reconstruction problem can be formulated as the estimation of the mean values of the mixture components and other mixture model parameters, given the 2D points describing the artery segmentations in the X-ray images.
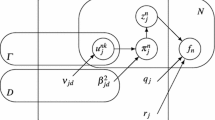
Let \(\varvec{X}^f = \{ \varvec{x}_n^f \in \mathbb {R}^2 \}_{n=1}^{N^f}\) be the set of 2D segmented artery points for the fth X-ray image of F retrospectively gated images. Similarly, let \(\varvec{Y} = \{ \varvec{y}_m \in \mathbb {R}^3 \}_{m=1}^{M}\) denotes the set of M 3D points corresponding to the mean values of the t-distributions. A multivariate t-distribution with mean \(\varvec{\mu }\), covariance \(\varvec{\varSigma }\) and degrees of freedom \(\nu \) can be written as an infinite mixture of scaled Gaussians
where \(\mathcal{{N}}(\cdot )\) and \(\mathcal{{G}}(\cdot )\) denote the Gaussian and Gamma distributions, respectively [6]. To compute the maximum-likelihood (ML) solution, u can be considered as an implicit latent variable introduced for each observation [6]. In our formulation, these latent variables are denoted by \(\varvec{U}^f = \{ u_n^f \in \mathbb {R} \}_{n=1}^{N^f}\) for the 2D points in fth X-ray image. Similarly, let \(\varvec{Z}^f = \{ \varvec{z}_n^f \in \mathbb {R}^{M} \}_{n=1}^{N^f}\) be the set of latent variables for the 2D points in fth X-ray image, where \(\varvec{z}_n^f\) is an M-dimensional binary vector that has only one non-zero entry. These vectors specify from which mixture component that the 2D segmentation point is generated. Finally, we can write the complete data probability for our mixture model as
where \(\mathbb {X} = \{ \varvec{X}^f \}_{f=1}^F\), \(\mathbb {Z} = \{ \varvec{Z}^f \}_{f=1}^F\) and \(\mathbb {U} = \{ \varvec{U}^f \}_{f=1}^F\). In addition, \(\mathcal{{H}}^f:\mathbb {R}^3 \rightarrow \mathbb {R}^2\) is the projection function for the fth frame, which is modelled by a weak-perspective or a perspective camera model. Furthermore, mixture model parameters are given by \(\varvec{\theta } = \big \{ \{ \pi _m \}_{m=1}^{M}, \varvec{Y}, \sigma ^2, \{ \nu _m \}_{m=1}^{M} \big \}\), where \(\pi _m\) and \(\nu _m\) are the mixing coefficient and degrees of freedom for mth component, respectively, and \(\sigma ^2\) is the isotropic variance for the mixture model components.
Given the 2D points segmented from X-ray images, \(\mathbb {X}\), the goal is to find the mixture model parameters that maximize the complete data log-likelihood function, i.e. \(\hat{\varvec{\theta }} = \hbox {arg max}_{\varvec{\theta }} \; \ln P(\mathbb {X}, \mathbb {Z}, \mathbb {U} | \varvec{\theta })\). The final 3D reconstruction of the coronary artery centreline points is given by the estimated mean values of the mixture model components, \(\hat{\varvec{Y}} \in \hat{\varvec{\theta }}\).
The ML estimation of the parameters \(\hat{\varvec{\theta }}\) can be found using expectation-conditional-maximization (ECM) algorithm [6]. In the E-step at iteration (t), we compute expectations \(E_{z_{nm}^{f}}(z_{nm}^{f} | \varvec{x}_n^f, \varvec{\theta }^{(t)}) = (\gamma _{nm}^f)^{(t)}\), \(E_{u_{n}^{f}}(u_{n}^{f} | \varvec{x}_n^f, \varvec{\theta }^{(t)}, z_{nm}^{f} = 1) = (\tau _{nm}^f)^{(t)}\) and \(E_{u_{n}^{f}}(\ln u_{n}^{f} | \varvec{x}_n^f, \varvec{\theta }^{(t)}, z_{nm}^{f} = 1)\). In the M-step, the parameters are updated from \(\varvec{\theta }^{(t)}\) to \(\varvec{\theta }^{(t+1)}\) using the expectations computed in E-step, such that the expectation of the log-likelihood function is maximized. After some algebra, it can be shown that the mean values of the t-distribution components can be computed by solving
For weak-perspective camera model, the Jacobian of the projection function is given by a linear projection operation which does not depend on \(\varvec{y}_m\). On the other hand, Jacobian is a function of \(\varvec{y}_m\) for the perspective camera model. Therefore, we obtain a closed form solution for weak-perspective model, and use numerical optimization for the perspective model to compute the reconstruction, which is defined by the mean values of the t-distribution components.
We obtain coronary artery centrelines from the set of reconstructed points by computing minimum spanning arborescence of a directed graph, whose vertices are the reconstructed points and edges are the possible connections between the neighbouring points. To this end, we select one of the reconstructed point as root of the graph based on our prior knowledge about the patient coordinate system. The graph is generated by connecting the root to all the remaining points, and each point to its neighbours inside 10 mm neighbourhood. The edge weights of the graph are determined by the Euclidean distance between the connecting points. After minimum spanning arborescence is computed using Edmonds algorithm [10], we apply some automatic pruning steps to obtain the final coronary arterial tree. First, we discard the reconstructed points that are located far from the remaining reconstructed points. Second, we remove the short branches that consist of less than 3 points. Finally, we fit a cubic spline to each branch of the coronary arterial tree (Fig. 1c and d).
3 Experiments and Results
Results on Synthetic Data: Due to the lack of ground truth information for validation, the quantitative validations of coronary artery reconstruction methods are typically based on synthetic data experiments. To this end, we generated two synthetic X-ray rotational angiography sequences and corresponding artery centreline segmentations using left coronary artery geometry of 4D XCAT phantom [9] (Fig. 1a). For both of the X-ray sequences, information related to acquisition, namely number of images (117), frame rate (30 fps), angular coverage (60\(^{\circ }\) RAO to 60\(^{\circ }\) LAO with 25\(^{\circ }\) CRA angulation), and the parameters defining the acquisition geometry model, was derived from a clinical dataset. In the first sequence, we employed the static geometry of the coronary arteries at end-diastole to generate the synthetic sequence. In the second sequence, we simulated cardiac motion where we set the heart beat rate to 70 beats per minute. Finally, we performed ECG gating on this sequence via a gating window of width \(10\,\%\) of the cardiac cycle. Effectively, a total of 11 images were selected that are close to end-diastolic phase, but still at different cardiac phases. For the experiments involving the static sequence, we selected the corresponding subset of X-ray images, which were acquired from the same viewpoints as the gated X-ray images from the dynamic sequence. We refer to the sets of X-ray images selected from the first and second sequence as StaticSet and CardiacSet, respectively.
In the synthetic data experiments, we assess: (i) the effect of erroneously segmented structures in the 2D segmentations (outliers), (ii) the effect of residual cardiac motion between the X-ray images, and (iii) the effect of number of 3D points describing the mixture model representation of the coronary arteries, on the coronary artery centreline reconstruction accuracy.
(a)–(d) Qualitative results on synthetic data with \(40\,\%\) outliers: (a) An example synthetic X-ray angiogram and the corresponding 2D segmentations (dark green) with outliers (light green) are shown. (b) The method is initialized using model points located on a regular spherical grid. (c) Reconstructed points are shown. (d) The final reconstruction output after pruning. In (b)–(d), the ground truth centrelines are shown in green, whereas reconstructed points or centrelines are shown in red. (e)–(f) Quantitative results with cardiac motion, different number of model components, and varying levels of outliers are presented using overlap and accuracy scores. (Color figure online)
To study the effect of outliers, we generated random 2D points and added these points to the corresponding 2D coronary artery segmentations. Specifically, we generated random 2D curves using a trajectory of a particle subject to Brownian motion, and sampled outlier points from these curves (Fig. 1a). The number of additional outlier points for each X-ray image was varied from \(0\,\%\) to \(40\,\%\) of the number of points in the 2D coronary artery segmentations for the same X-ray image with \(10\,\%\) increments. This procedure is carried out for all X-ray images in StaticSet and CardiacSet, so that we can additionally study the effect of residual motion. To initialize both the number and the spatial locations of the mixture model components, we assumed that points are located on a regular grid in spherical coordinates centred at the origin of the patient coordinate system. By changing the radial sampling rate of the grid, we adjusted M to 168 and 210 points (Fig. 1b).
In all of the experiments, the reconstruction accuracy measured in terms of two 3D centreline to centreline scores, namely overlap (OV\(_{3D}\)) and accuracy (AC\(_{3D}\)) scores, which were introduced as a part of the standardized evaluation framework described in [7]. Briefly, the overlap score measures how the reconstructed centreline agrees with the ground truth, based on the labelling of correspondences between reconstructed points and the ground truth centreline points as false positive, negative and true positive. On the other hand, the accuracy score measures the mean 3D Euclidean distance between the true positive correspondences.
We repeated the reconstruction experiments 15 times with different random outlier points in each outlier level. The qualitative and quantitative results of the synthetic data experiments are shown in Fig. 1. Results indicate that the performance of our method does not decrease drastically, even in the presence of high level of outlier points. As expected, the scores for CardiacSet, generated under cardiac motion, are lower, but comparable to the results from StaticSet.
As shown in Fig. 1c, the reconstructed points may contain some scattered points as we increase the outlier level. This can be explained as follows: when there is a significant number of outliers, the possibility of consistent point correspondences between various projection frames increases. As a result, some mixture model components are displaced towards those outliers. However, with lower outlier level, the heavy tails of t-distribution mixture components sufficiently explain those points. Therefore, no significant displacement of the mixture components is observed. Additionally, the situation is also aggravated when we increase the number of mixture model components as reflected by the increased dispersion of the overlap scores.
Results on clinical data: (a) Projection of the reconstructed coronary arteries (red) is overlaid on top of the segmented coronary arteries (green) on the left, and the corresponding 3D reconstruction result is shown on the right. (b) The results of the proposed algorithm for both synthetic and clinical experiments are compared with the relevant literature. These results were obtained using different synthetic and clinical datasets, but nonetheless show the accuracies of the various techniques. (Color figure online)
Results on Clinical Data: We reconstructed centrelines from 2 RCA and 1 LCA rotational angiography studies. The angular coverage of the acquisitions was 120\(^{\circ }\) (60\(^{\circ }\) RAO to 60\(^{\circ }\) LAO with 25\(^{\circ }\) CRA/CAU angulation). From each study, 4 images (one from each cardiac cycle) were selected via ECG gating.
Coronary artery segmentations from X-ray angiograms are carried out by the automatic algorithm given in [8]. Because the ground truth is not available for the clinical data, we present qualitative 3D reconstruction result and its back-projection onto the X-ray images (Fig. 2a). These results underscore the robustness of the algorithm in the presence of outliers. Although using TMM makes our reconstruction robust to outliers, reconstructed centrelines can be further improved if better segmentations (free from obvious errors or missing parts) are provided. To this end, we segmented centrelines using a workflow similar to the one proposed in [5], and cleaned the resulting segmentations. To facilitate quantitative evaluation, we computed 2D reprojection error (RPE\(_{2D}\)), which is defined as the average 2D Euclidean distance between 2D ground truth points and 2D projections of the reconstructed centrelines. The results are shown in Fig. 2b in comparison with the results from the relevant literature.
There is a trade-off between target accuracy (associated with M), and the computation time. Mean computation time for the clinical dataset was 21.8 min with \(M=168\) points describing the arteries.
4 Conclusion
In this paper, we propose a novel probabilistic framework to reconstruct coronary artery centrelines from retrospectively gated X-ray angiography images using a mixture of Student’s t distributions. Given 2D segmentations of coronary arteries from X-ray images, we formulate the reconstruction problem as ML estimation of mixture model parameters. The framework is highly versatile and does not require point correspondences across various projections. The heavy tail of Student’s t-distribution allows robust reconstruction and good handling of outliers. The quantitative validation on synthetic data indicates that our method can cope with reasonable level of erroneously segmented parts in the 2D coronary artery segmentations. As demonstrated in the experiments using clinical X-ray angiography images, this aspect enables using coronary segmentation algorithms which may produce some level of outliers. Proposed method provides a convenient framework to incorporate prior information (e.g. Bayesian priors enforcing linearity of local structures, and sparsity of mixture components), owing to its probabilistic formulation. This topic will be the focus of our future research.
References
Çimen, S., Gooya, A., Grass, M., Frangi, A.F.: Reconstruction of coronary arteries from X-ray angiography: a review. Med. Image Anal. 32, 46–68 (2016)
Cong, W., Yang, J., Ai, D., Chen, Y., Liu, Y., Wang, Y.: Quantitative analysis of deformable model-based 3-D reconstruction of coronary artery from multiple angiograms. IEEE Trans. Biomed. Eng. 62(8), 2079–2090 (2015)
Hartley, R., Zisserman, A.: Multiple View Geometry in Computer Vision, 2nd edn. Cambridge University Press, Cambridge (2004)
Jandt, U., Schäfer, D., Grass, M., Rasche, V.: Automatic generation of 3D coronary artery centerlines using rotational X-ray angiography. Med. Image Anal. 13(6), 846–858 (2009)
Liao, R., Luc, D., Sun, Y., Kirchberg, K.: 3-D reconstruction of the coronary artery tree from multiple views of a rotational X-ray angiography. Int. J. Cardiovasc. Imaging 26(7), 733–749 (2010)
Peel, D., McLachlan, G.J.: Robust mixture modelling using the t distribution. Stat. Comput. 10(4), 339–348 (2000)
Schaap, M., Metz, C.T., van Walsum, T., van der Giessen, A.G., Weustink, A.C., Mollet, N.R., Bauer, C., Bogunović, H., Castro, C., Deng, X., Dikici, E., O’Donnell, T., Frenay, M., Friman, O., Hernández Hoyos, M., Kitslaar, P.H., Krissian, K., Kühnel, C., Luengo-Oroz, M.A., Orkisz, M., Smedby, O., Styner, M., Szymczak, A., Tek, H., Wang, C., Warfield, S.K., Zambal, S., Zhang, Y., Krestin, G.P., Niessen, W.J.: Standardized evaluation methodology and reference database for evaluating coronary artery centerline extraction algorithms. Med. Image Anal. 13(5), 701–714 (2009)
Schneider, M., Sundar, H.: Automatic global vessel segmentation and catheter removal using local geometry information and vector field integration. In: Proceedings IEEE ISBI, pp. 45–48. IEEE (2010)
Segars, W.P., Sturgeon, G., Mendonca, S., Grimes, J., Tsui, B.M.W.: 4D XCAT phantom for multimodality imaging research. Med. Phys. 37(9), 4902–4915 (2010)
Tarjan, R.E.: Finding optimum branchings. Networks 7(1), 25–35 (1977)
Yang, J., Cong, W., Chen, Y., Fan, J., Liu, Y., Wang, Y.: External force back-projective composition and globally deformable optimization for 3-D coronary artery reconstruction. Phys. Med. Biol. 59(4), 975–1003 (2014)
Acknowledgements
This project was partly supported by the Marie Curie Individual Fellowship (625745, A. Gooya).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Çimen, S., Gooya, A., Ravikumar, N., Taylor, Z.A., Frangi, A.F. (2016). Reconstruction of Coronary Artery Centrelines from X-Ray Angiography Using a Mixture of Student’s t-Distributions. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9902. Springer, Cham. https://doi.org/10.1007/978-3-319-46726-9_34
Download citation
DOI: https://doi.org/10.1007/978-3-319-46726-9_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46725-2
Online ISBN: 978-3-319-46726-9
eBook Packages: Computer ScienceComputer Science (R0)