Abstract
We present a novel methodology for the automated detection of breast lesions from dynamic contrast-enhanced magnetic resonance volumes (DCE-MRI). Our method, based on deep reinforcement learning, significantly reduces the inference time for lesion detection compared to an exhaustive search, while retaining state-of-art accuracy.
This speed-up is achieved via an attention mechanism that progressively focuses the search for a lesion (or lesions) on the appropriate region(s) of the input volume. The attention mechanism is implemented by training an artificial agent to learn a search policy, which is then exploited during inference. Specifically, we extend the deep Q-network approach, previously demonstrated on simpler problems such as anatomical landmark detection, in order to detect lesions that have a significant variation in shape, appearance, location and size. We demonstrate our results on a dataset containing 117 DCE-MRI volumes, validating run-time and accuracy of lesion detection.
Supported by Australian Research Council through grants DP140102794, CE140100016 and FL130100102.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Breast cancer is amongst the most commonly diagnosed cancers in women [1, 2]. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) represents one of the most effective imaging techniques for monitoring younger, high-risk women, who typically have dense breasts that show poor contrast in mammography [3]. DCE-MRI is also useful during surgical planning once a suspicious lesion is found on a mammogram [3]. The first stage in the analysis of these 4D (3D over time) DCE-MRI volumes consists of the localisation of breast lesions. This is a challenging task given the high dimensionality the data (4 volumes each containing \(512 \times 512 \times 128\) voxels), the low signal to noise ratio of the dynamic sequence and the variable size and shape of breast lesions (see Fig. 1). Therefore, a computer-aided detection (CAD) system that automatically localises breast lesions in DCE-MRI data would be a useful tool to facilitate radiologists. However, the high dimensionality of DCE-MRI requires computationally efficient methods for lesion detection to be developed to be viable for practical use.
Current approaches to lesion detection in DCE-MRI rely on extracting hand-crafted features [4, 5] and exhaustive search mechanisms [4,5,6] in order to handle the variability in lesion appearance, shape, location and size. These methods are both computationally complex and potentially sub-optimal, resulting in false alarms and missed detections. Similar issues in the detection of visual objects have motivated the computer vision community to develop efficient detectors [7, 8], like the Faster R-CNN [7]. However, these models need large annotated training sets making their application in medical image analysis (MIA) challenging [9]. Alternatively, Caicedo and Lazebnik [8] have recently proposed the use of a deep Q-network (DQN) [10] for efficient object detection that allows us to deal with the limited amount of data. Its adaptation to MIA applications has to overcome two additional obstacles: (1) the extension from visual object classes (e.g., animals, cars, etc.) to objects in medical images, such as tumours, which tend to have weaker consistency in terms of shape, appearance, location, background and size; and (2) the high dimensionality of medical images, which presents practical challenges with respect to the DQN training process [10]. Ghesu et al. [11] have recently adapted DQN [10] to anatomical landmark detection, but did not address the obstacles mentioned above because the visual classes used in their work have consistent patterns and are extracted from fixed small-size regions of the medical images.
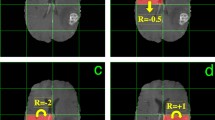
Here, we introduce a novel algorithm for breast lesion detection from DCE-MRI inspired by a previously proposed DQN [8, 10]. Our main goal is the reduction of run time complexity without a reduction in detection accuracy. The proposed approach comprises an artificial agent that automatically learns a policy, describing how to iteratively modify the focus of attention (via translation and scale) from an initial large bounding box to a smaller bounding box containing a lesion, if it exists (see Fig. 2). To this end, the agent constructs a deep learning feature representation of the current bounding box, which is used by the DQN to decide on the next action, i.e., either to translate or scale the current bounding box or to trigger the end of the search process. Our methodology is the first DQN [10] that can detect such visually challenging objects. In addition, unlike [11] that uses a fixed small-size bounding box, our DQN utilises a variable-size bounding box. We evaluate our methodology on a dataset of 117 patients (58 for training and 59 for testing). Results show that our methodology achieves a similar detection accuracy compared to the state of the art [5, 6], but with significantly reduced run times.
2 Literature Review
Automated approaches for breast lesion detection from DCE-MRI are typically based on exhaustive search methods and hand-designed features [4, 5, 12, 13]. Vignati et al. [12] proposed a method that thresholds an intensity normalised DCE-MRI to detect voxel candidates that are merged to form lesion candidates, from which hand-designed region and kinetic features are used in the classification process. As shown in Table 1, this method has low accuracy that can be explained by the fact that this method makes strong assumptions about the role of DCE-MRI intensity and does not utilise texture, shape, location and size features. Renz et al. [13] extended Vignati et al.’s work [12] with the use of additional hand-designed morphological and dynamical features, showing more competitive results (see Table 1). Further improvements were obtained by Gubern-Merida et al. [4], with the addition of shape and appearance hand-designed features, as shown in Table 1. The run-time complexity of the approaches above can be summarised by the mean running time (per volume) shown by Vignati et al.’s work [12] in Table 1, which is likely the most efficient of these three approaches [4, 12, 13]. McClymont et al. [5] extended the methods above with the unsupervised voxel clustering for the initial detection of lesion candidates, followed by a structured output learning approach that detects and segments lesions simultaneously. This approach significantly improves the detection accuracy, but at a substantial increase in computational cost (see Table 1). The multi-scale deep learning cascade approach [6] reduced the run-time complexity, allowed the extraction of optimal and efficient features, and had a competitive detection accuracy as shown in Table 1.
There are two important issues regarding previously proposed approaches: the absence of a common dataset to evaluate different methodologies and the lack of a consistent lesion detection criterion. Whereas detections in [12, 13] were visually inspected by a radiologist, [4, 5] considered a lesion detected if a (single) voxel in the ground truth was detected. In [6] a more precise criterion (minimum Dice coefficient of 0.2 between ground truth and candidate bounding box) was used - in the experiment, we adopt this Dice > 0.2 criterion and use the same dataset as a few previous studies [5, 6].
3 Methodology
In this section, we first define the dataset, then the training and inference stages of our proposed methodology, shown in Fig. 2.
3.1 Dataset
The data is represented by a set of 3D breast scans \(\mathcal{D} = \left\{ \left( \mathbf {x}, \mathbf {t}, \{ \mathbf {s}^{(j)} \}_{j = 1}^M \right) _i \right\} _{i=1}^{|\mathcal{D}|}\), where each, \(\mathbf {x},\mathbf {t}:\varOmega \rightarrow \mathbb R\) denotes the first DCE-MRI subtraction volume and the T1-weighted anatomical volume, respectively, with \(\varOmega \in \mathbb R^3\) representing the volume lattice of size \({w \times h \times d}\); \(\mathbf {s}^{(j)} : \varOmega \rightarrow \{0,1\}\) represents the annotation for the \(j^{\text{ th }}\) lesion present, with \(\mathbf {s}^{(j)}(\omega ) = 1\) indicating presence of lesion at voxel \(\omega \in \varOmega \). The entire dataset is patient-wise split such that the mutually exclusive training and testing datasets are represented by \(\mathcal {T},\mathcal {U} \subset \mathcal {D}\), where \(\mathcal {T} \bigcup \mathcal {U} = \mathcal {D}\).
3.2 Training
The proposed DQN [10] model is trained via interactions with the DCE-MRI dataset through a sequence of observations, actions and rewards. Each observation is represented by \(\mathbf {o} = f(\mathbf {x}({\mathbf {b}}))\), where \(\mathbf {b}=[b_x,b_y,b_z,b_w,b_h,b_d] \in \mathbb R^6\) (where \(b_x,b_y,b_z\) represent the top-left-front corner and \(b_w,b_h,b_d\) denotes the lower-right-back corner of the bounding box) indexes the input DCE-MRI data \(\mathbf {x}\), and f(.) denotes a deep residual network (ResNet) [14, 15], defined below. Each action is denoted by \(a \in \mathcal {A} = \{ l_x^+,l_x^-,l_y^+,l_y^-,l_z^+,l_z^-,s^+,s^-,w \}\), where l, s, w represent translation, scale and trigger actions, with the subscripts x, y, z denoting the horizontal, vertical or depth translation, and superscripts \(+,-\) meaning positive or negative translation and up or down scaling. The reward when the agent chooses the action \(a=w\) to move from \(\mathbf {o}\) to \(\mathbf {o}'\) is defined by:
where d(.) is the Dice coefficient between a map formed by the bounding box \(\mathbf {o} = f(\mathbf {x}({\mathbf {b}}))\) and the segmentation map \(\mathbf {s}\), \(\eta =10\) and \(\tau _w=0.2\) (these values have been empirically defined - for instance, we found that increasing \(\eta \) to 10.0 from 3.0 used in [8] helped triggering when finding a lesion). For the remaining of the actions in \(\mathcal {A} {\setminus } \{w\}\), the rewards are defined by:
The training process models a DQN that maximises cumulative future rewards with the approximation of the following action-value function: \(Q^*(\mathbf {o},a) = \max _{\pi } \mathbb E [ r_t + \gamma r_{t+1} + \gamma ^2 r_{t+2} + ... \; | \;\mathbf {o}_t = \mathbf {o}, a_t = a, \pi ],\) where \(r_t\) denotes the reward at time step t, \(\gamma \) represents a discount factor per time step, and \(\pi \) is the behaviour policy. This action-value function is modelled by a DQN \(Q(\mathbf {o},a, \theta )\), where \(\theta \) denotes the network weights. The training of \(Q(\mathbf {o},a, \theta )\) is based on experience replay memory and the target network [10]. Experience replay uses a dataset \(\mathcal {E}_t = \{ e_1,...,e_t \}\) built with the agent’s experiences \(e_t = (\mathbf {o}_t,a_t,r_t,\mathbf {o}_{t + 1})\), and the target network with parameters \(\theta _i^-\) computes the target values for the DQN updates, where the values \(\theta _i^-\) are held fixed and updated periodically. The loss function for modelling \(Q(\mathbf {o},a, \theta )\) minimises the mean-squared error of the Bellman equation, as in:
In the training process, we follow an \(\epsilon \)-greedy strategy to balance exploration and exploitation: with probability \(\epsilon \), the agent explores, and with probability 1-\(\epsilon \), it will follow the current policy \(\pi \) (exploitation) for training time step t. At the beginning of the training, we set \(\epsilon =1\) (i.e., pure exploration), and decrease \(\epsilon \) as the training progresses (i.e., increase exploitation). Furthermore, we follow a modified guided exploration: with probability \(\kappa \), the agent will select a random action and with probability \(1-\kappa \), it will select an action that produces a positive reward. This modifies the guided exploration in [8] by adding randomness to the process, aiming to improve generalisation. Finally, the ResNet [14, 15], which produces the observation \(\mathbf {o} = \mathbf {x}({\mathbf {b}})\), is trained to decide whether a random bounding box \(\mathbf {b}\) contains a lesion. A training sample is labelled as positive if \(d(\mathbf {o}, \mathbf {s}_j ) \ge \tau _w\), and negative, otherwise. It is important to notice that this way of labelling random training samples can provide a large and balanced training set, extracted at several locations and scales, that is essential to train the large capacity ResNet [14, 15]. In addition, this way of representing the bounding box means that we are able to process varying-size input bounding box, which is an advantage compared to [11].
3.3 Inference
The trained DQN model is parameterised by \(\theta ^*\) learned in (3) and is defined by a multi-layer perceptron [8] that outputs the action-value function for the observation \(\mathbf {o}\). The action to follow from the current observation is defined by:
Finally, given that the number and location of lesions are unknown in a test DCE-MRI, this inference is initialised with different bounding boxes at several locations, and it runs until it either finds the lesion (with the selection of the trigger action), or runs for a maximum number of 20 steps.
4 Experiments
The database used to assess our proposed methodology contains DCE-MRI and T1-weighted anatomical datasets from 117 patients [5]. For the DCE-MRI, the first volume was acquired before contrast agent injection (pre-contrast), and the remaining volumes were acquired after contrast agent injection. Here we use only one volume represented by the first subtraction from DCE-MRI: the first post-contrast volume minus pre-contrast volume. The T1-weighted anatomical is used only to extract the breast region from the initial volume [5], as a pre-processing stage. The training set contains 58 patients annotated with 72 lesions, and the testing set has 59 patients and 69 lesions to allow a fair comparison with [6]. The detection accuracy is assessed by the proportion of true positives (TPR) detected in the training and testing sets as a function of the number of false positives per image (FPI), where a candidate lesion is assumed to be a true positive if the Dice coefficient between the candidate lesion bounding box and the ground truth annotation bounding box is at least 0.2 [16]. We also measure the running time of the detection process using the following computer: CPU: Intel Core i7 with 12 GB of RAM and a GPU Nvidia Titan X 12 GB.
The pre-processing stage of our methodology consists of the extraction of each breast region (from T1-weighted) [5], and separate each breast into a resized volume of (100 \(\times \) 100 \(\times \) 50) voxels. For training, we select breast region volumes that contain at least one lesion, but if a breast volume has more than one lesion, one of them is randomly selected to train the agent. For testing, a breast may contain none, one or multiple lesions. The observation \(\mathbf {o}\) used by DQN is produced by a ResNet [14] containing five residual blocks. The input to the ResNet is fixed at (100 \(\times \) 100 \(\times \) 50) voxels. We extract 16 K patches (8 K positives and 8 K negatives) from the training set to train the ResNet to classify a bounding box as positive or negative for a lesion, where a bounding box is labelled as positive if the Dice coefficient between the lesion candidate and the ground truth annotation is at least 0.6. This ResNet provides a fixed size representation for \(\mathbf {o}\) of size 2304 (extracted before the last convolutional layer).
The DQN is represented by a multilayer perceptron with two layers, each containing 512 nodes, that outputs nine actions: six translations (by one third of the size of the corresponding dimension), two scales (by one sixth in all dimensions) and a trigger (see Sect. 3.2). For training this DQN, the agent starts an episode with a centred bounding box occupying 75% of the breast region volume. The experience replay memory \(\mathcal {E}\) contains 10 K experiences, from which 100 mini-batch samples are drawn to minimise the loss (3). The DQN is trained with Adam, using a learning rate of \(1 \times 10^{-6}\), and the target network is updated after running one episode per volume of the training set. For the \(\epsilon \)-greedy strategy (Sect. 3.2), \(\epsilon \) decreases linearly from 1 to 0.1 in 300 epochs, and during exploration,the balance between random exploration and modified guided exploration is given by \(\kappa = 0.5\). During inference, the agent follows the policy in (4), where for every breast region volume, it starts at a centred bounding box that covers 75% of the volume. Then it starts at each of the eight non-overlapping (50, 50, 25) bounding boxes corresponding to each of the corners. Finally, it is initialised at another four (50, 50, 25) bounding boxes centred at the intersections of the previous bounding boxes. The agent is allowed a maximum number of 20 steps to trigger, otherwise, no lesion is detected.
4.1 Results
We compare the training and testing results of our proposed DQN with the multi-scale cascade [6] and structured output approach [5] in the table in Fig. 3. In addition, we show the Free Response Operating Characteristic (FROC) curve in Fig. 3 comparing our approach (using a varying number of initialisations that lead to different TPR and FPI values) with the multi-scale cascade [6]. Finally, we show examples of the detections produced by our method in Fig. 4.
FROC curve showing TPR vs FPI and run times for DQN and the multi-scale cascade [6] (left) TPR, FPR and mean inference time per case (i.e. per patient) for each method (right). Note run time for Ms-C is constant over the FPI range.
We use a paired t-test to estimate the significance of the inference times between our approach and the multi-scale cascade [6], giving \(p \le 9 \times 10^{-5}\).
5 Discussion and Conclusion
We have presented a DQN method for lesion detection from DCE-MRI that shows similar accuracy to state of the art approaches, but with significantly reducing detection times. Given that we did not attempt any code optimisation, we believe that the run times have the potential for further improvement. For example, inference uses several initialisations (up to 13), which could be run in parallel as they are independent, decreasing detection time by a factor of 10. The main bottleneck of our approach is the volume resizing stage that transforms the current bounding box to fit the ResNet input - currently representing 90% of the inference time. A limitation of this work is that we do not have an action to change the aspect ratio of the bounding box, which may improve detection of small elongated lesions. Finally, during training, we noted that the most important parameter to achieve good generalisation is the balance between exploration and exploitation. We observed that the best generalisation was achieved when \(\epsilon = 0.5\) (i.e. half of the actions correspond to exploration and half to exploitation of the current policy). Future research will improve run-time performance via learning smarter search strategies. For instance, we would like to avoid re-visiting regions that have already been determined to be free from lesions with high probability. At present we rely on the training data to discourage such moves, but there may be more explicit constraints to explore. We would like to acknowledge NVIDIA for providing the GPU used in this work.
References
Smith, R.A., Andrews, K., Brooks, D., et al.: Cancer screening in the United States, 2016: a review of current American cancer society guidelines and current issues in cancer screening. CA Cancer J. Clin. 66, 96–114 (2016)
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics, 2016. CA Cancer J. Clin. 66(1), 7–30 (2016)
Siu, A.L.: Screening for breast cancer: US preventive services task force recommendation statement. Ann. Intern. Med. 164, 279–296 (2016)
Gubern-Mérida, A., Martí, R., Melendez, J., et al.: Automated localization of breast cancer in DCE-MRI. Med. Image Anal. 20(1), 265–274 (2015)
McClymont, D., Mehnert, A., Trakic, A., et al.: Fully automatic lesion segmentation in breast MRI using mean-shift and graph-cuts on a region adjacency graph. JMRI 39(4), 795–804 (2014)
Maicas, G., Carneiro, G., Bradley, A.P.: Globally optimal breast mass segmentation from DCE-MRI using deep semantic segmentation as shape prior. In: 14th International Symposium on Biomedical Imaging (ISBI), pp. 305–309. IEEE (2017)
Ren, S., He, K., Girshick, R., Sun, J.: Faster R-CNN: towards real-time object detection with region proposal networks. In: NIPS, pp. 91–99 (2015)
Caicedo, J.C., Lazebnik, S.: Active object localization with deep reinforcement learning. In: CVPR, pp. 2488–2496 (2015)
Akselrod-Ballin, A., Karlinsky, L., Alpert, S., Hasoul, S., Ben-Ari, R., Barkan, E.: A region based convolutional network for tumor detection and classification in breast mammography. In: Carneiro, G., et al. (eds.) LABELS/DLMIA -2016. LNCS, vol. 10008, pp. 197–205. Springer, Cham (2016). doi:10.1007/978-3-319-46976-8_21
Mnih, V., Kavukcuoglu, K., Silver, D., et al.: Human-level control through deep reinforcement learning. Nature 518(7540), 529–533 (2015)
Ghesu, F.C., Georgescu, B., Mansi, T., Neumann, D., Hornegger, J., Comaniciu, D.: An artificial agent for anatomical landmark detection in medical images. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9902, pp. 229–237. Springer, Cham (2016). doi:10.1007/978-3-319-46726-9_27
Vignati, A., Giannini, V., De Luca, M., et al.: Performance of a fully automatic lesion detection system for breast DCE-MRI. JMRI 34(6), 1341–1351 (2011)
Renz, D.M., Böttcher, J., Diekmann, F., et al.: Detection and classification of contrast-enhancing masses by a fully automatic computer-assisted diagnosis system for breast MRI. JMRI 35(5), 1077–1088 (2012)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016)
Huang, G., Sun, Y., Liu, Z., Sedra, D., Weinberger, K.Q.: Deep networks with stochastic depth. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9908, pp. 646–661. Springer, Cham (2016). doi:10.1007/978-3-319-46493-0_39
Dhungel, N., Carneiro, G., Bradley, A.P.: Automated mass detection in mammograms using cascaded deep learning and random forests. In: DICTA. IEEE (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Maicas, G., Carneiro, G., Bradley, A.P., Nascimento, J.C., Reid, I. (2017). Deep Reinforcement Learning for Active Breast Lesion Detection from DCE-MRI. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10435. Springer, Cham. https://doi.org/10.1007/978-3-319-66179-7_76
Download citation
DOI: https://doi.org/10.1007/978-3-319-66179-7_76
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66178-0
Online ISBN: 978-3-319-66179-7
eBook Packages: Computer ScienceComputer Science (R0)