Abstract
The trend towards large-scale studies including population imaging poses new challenges in terms of quality control (QC). This is a particular issue when automatic processing tools such as image segmentation methods are employed to derive quantitative measures or biomarkers for further analyses. Manual inspection and visual QC of each segmentation result is not feasible at large scale. However, it is important to be able to detect when an automatic method fails to avoid inclusion of wrong measurements into subsequent analyses which could otherwise lead to incorrect conclusions. To overcome this challenge, we explore an approach for predicting segmentation quality based on reverse classification accuracy, which enables us to discriminate between successful and failed cases. We validate this approach on a large cohort of cardiac MRI for which manual QC scores were available. Our results on 7,425 cases demonstrate the potential for fully automatic QC in the context of large-scale population imaging such as the UK Biobank Imaging Study.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Biomedical image data are increasingly processed with automated image analysis pipelines which employ a variety of tools to extract clinically useful information. It is important to understand the limitations of such pipelines and assess the quality of the results being reported. This is a particular issue when we consider large-scale population imaging databases comprising thousands of images, e.g., as done in the UK Biobank (UKBB) Imaging Study [1]. There are often many modules in automated pipelines [2] where each may contribute to inaccuracies in the final output and reduce the overall quality of the analysis, e.g., intensity normalisation, segmentation, registration and feature extraction. On a large scale, it is infeasible to perform a manual, visual inspection of all outputs, and even more difficult to perform quality control (QC) within the pipeline itself. We break down this challenge and focus on the automated QC of image segmentation. Many segmentation methods have been developed, from graph cuts to advanced algorithms involving machine learning [3]. Their performance is traditionally evaluated on a set of labelled data using an evaluation metric which computes similarity between the prediction and a reference ‘ground truth’ (GT). Popular metrics include volumetric overlap [4], surface distances or other statistical measures [5]. Due to the lack of actual GT, manual expert annotations are used as reference, despite inter- and intra-rater variability. But such reference segmentations are not available once a segmentation method is deployed in a clinical practice. Evaluating the expected average performance of an algorithm is less important as being able to assess the quality on a per-case basis, and it is crucial to identify cases where the segmentation has failed. We show that we can effectively predict the per-case quality of 3D cardiac MRI (CMR) segmentations which enables fully automated QC in a large-scale population studies.
Related work. We adopt the recently proposed approach of reverse classification accuracy (RCA) [6]. RCA is a general framework for predicting the performance of a deployed segmentation method on a case-by-case basis when the GT is unknown. This makes it stand out from other evaluation frameworks such as reverse validation and reverse testing [7, 8], which only provide an estimate of the average performance. In contrast to other approaches for predicting segmentation quality [9], RCA has the advantage of not requiring a training database with good and bad segmentations. Image quality assessment (IQA) has previously been studied on the dataset of 5,000 UKBB cardiac cineMRI [10]. The value here is in the cardiac specific ontology and derived rules could be used to reduce variability in qualitative IQA terminology. However, the rudimentary numerical analysis of segmentations, a score in the range \([1 \ 3]\), does not utilise image data, nor provide a quantitative evaluation using traditional metrics on a per-case basis. Further work on automating CMR IQA includes a field-of-view assessment algorithm [11]. Here, Convolutional Neural Networks were used to identify missing apical and basal slices in a test set after training on the UKBB pilot database. A previous study on an unsupervised approach to artefact detection [10] employs dictionary learning to annotate affected images, but appears insensitive to artefacts caused by movement. These approaches are not directly applicable to predicting segmentation quality on a per-case basis.
Contributions are two-fold: (1) a thorough validation of RCA for the application of cardiac MR segmentation. Our results indicate highly accurate predictions of segmentation quality across various metrics; (2) Feasibility study of using RCA for automatic QC in large-scale studies. RCA predictions correlate with manual QC scores and enable outlier detection in a large set of 7,425 cases.
2 Method and Material
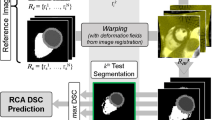
In RCA the idea is to build a classifier based on the test image using its predicted segmentation as pseudo GT. Assuming that the predicted segmentation is of good quality, the classifier should be able to segment at least one reference image with high accuracy. This is a fair assumption in the context of machine learning, where it is generally assumed that training data is similar to test data. The advantage of RCA is that it detects whether there is a mismatch between training and test data. If the predicted segmentation is of poor quality, the RCA classifier should fail on all reference images. The performance of the RCA classifier on the reference set is measured with a chosen quality metric (e.g., Dice similarity coefficient (DSC)), and the highest score determines the quality estimate for the test segmentation. The RCA classifier could be an Atlas Forest [12] or, as used in this paper, a registration algorithm [13]. Figure 1 gives an overview of RCA as applied in our study. More details can be found in [6].
We validate RCA for the purpose of predicting cardiac image segmentation using carefully segmented atlases. Each atlas contains a short-axis end-diastolic 3D MRI, its manual segmentation and 6 anatomical landmarks. The images have a pixel-resolution of \(1.25 \times 1.25 \times 2.0\) mm and span \(256 \times 256 \times 56\) voxels. Landmarks are located at the apex, base and four extremes of the left-ventricular (LV) cavity in the central slice. Each manual labelmap identifies voxels belonging to the LV cavity (class 1), LV myocardium (class 2) and right-ventricular (RV) cavity (class 4). These separate the heart from the background class, labelled 0. Class 3 corresponds to papillary muscles, not considered in this paper.
Overview of the RCA framework for 3D cardiac MR with single-atlas registration. In practice, the ground truth (GT) of the test image is unavailable so ‘real DSC’ is unknown. Reference images with available GT are segmented using the prediction on the test image. The best score obtained over all references is used as a proxy for determining the quality of the prediction on the test image.
For the RCA reference images, we use a set \( \mathbf {R_{\mathrm {I}}} = \left\{ R_{\mathrm {I}}^{1}, \cdots , R_{\mathrm {I}}^{n} \right\} \) of \(n = 20\) cardiac atlases with reference segmentations \( \mathbf {R_{\mathrm {G}}} = \left\{ R_{\mathrm {G}}^{1}, \cdots , R_{\mathrm {G}}^{n} \right\} \). For validation, we use set \( \mathbf {T_{\mathrm {I}}} = \left\{ T_{\mathrm {I}}^{1}, \cdots , T_{\mathrm {I}}^{m} \right\} \) of another \( m = 80\) atlases with reference segmentations \( \mathbf {T_{\mathrm {G}}} = \left\{ T_{\mathrm {G}}^{1}, \cdots , T_{\mathrm {G}}^{m} \right\} \). We then employ an automatic segmentation method to obtain predicted segmentations \(\mathbf {T_{S}} = \left\{ T_{\mathrm {S}}^{1}, \cdots , T_{\mathrm {S}}^{m} \right\} \) for each image in \(\mathbf {T_{\mathrm {I}}}\). Using RCA we estimate the quality of those predicted segmentations and compare the estimates to the real quality w.r.t. reference GT.
In order to validate that RCA is able to accurately predict segmentation performance, we generate automatic segmentations with varying quality. To this end, we employ Random Forests (RFs) with \(T=500\) and a maximum depth of \(D=40\) trained on the set of 20 reference images \(\{ \mathbf {R_{\mathrm {I}}}, \mathbf {R_{\mathrm {G}}} \}\). RFs allow us to produce a variety of test segmentations with degraded segmentation quality by limiting the depth of the trees during test time. We obtain sets of 80 ‘poor’, ‘average’ and ‘good’ segmentations by using depths of 5, 10 and 40 respectively. Thus, a total of 240 segmentations are used in our validation study.
To apply RCA, all reference images and their segmentations \(\mathbf {\lbrace R_{\mathrm {I}}, R_{\mathrm {G}} \rbrace }\) are first registered to the ith test image \(T_{\mathrm {I}}^{i}\) by performing a rigid registration. We then perform non-linear registration of \(T_{\mathrm {I}}^{i}\) to each reference image in \(\mathbf {R_{\mathrm {I}}}\) to get warped segmentations \(\mathbf {W_{\mathrm {S}}^{i}}\). For each segmentation pair in \(\lbrace \mathbf {R_{\mathrm {G}}},\mathbf {W_{\mathrm {S}}^{i}} \rbrace \) we evaluate DSC, mean surface distance (MSD), root-mean-square error (RMS) and Hausdorff distance (HD). The smallest of these values (or the largest in the case of DSC) over all pairs is taken to be the predictor of segmentation accuracy for \(T_{\mathrm {S}}^{i}\). For each test image, we report the evaluation metrics for each class label: 1, 2 and 4. We also report the average of each metric by evaluating the segmentation as a whole rather than calculating a mean across the classes.
To identify individual cases where segmentation has failed, we incorporate a classification strategy similar to [6]. We consider a 2-group classification where DSC scores in the range [0.0 0.7) are considered ‘poor’ and in range [0.7 1.0] are ‘good’. These boundaries are somewhat arbitrary and would be adjusted for a particular use-case. Other strategies could be employed on a task-specific basis, e.g., formulation as outlier detection with further statistical measures.
3 Results
We perform RCA validation on all 240 3D segmentations using a reference set of 20 atlases. A summary of the results is shown in Table 1. We obtain low mean absolute error (MAE) across all evaluation metrics and all class labels. The scatter plots in Fig. 2 on real and predicted scores illustrate the very good performance of RCA in predicting segmentation scores. We also find that from the 240 test segmentations, RCA is able to classify ‘good’ (\(\mathrm {DSC} \in \left[ 0.7 \ 1.0\right] \)) and ‘poor’ (\(\mathrm {DSC} \in [0.1 \ 0.7)\)) segmentations with an accuracy of 83%. For all evaluation metrics, there is a strong, positive linear relationship between predicted and real values with \(r \in \left[ 0.86 \ 0.94 \right] \) and \(p < 0.001\). This analysis shows RCA’s ability to correctly identify segmentations of poor quality - one of the primary motivations of this work. When employed in an automated QC framework, we would be less interested in the actual quality scores but would hope to be able to flag up failed cases and select high quality results for further analyses. The threshold could be chosen depending on the application’s requirements for what qualifies as a ‘good’ segmentation. Failed segmentations could be re-segmented with different parameters, regenerated with alternative methods, sent to a user for manual inspection, or discarded from further analyses. The RCA validation process was carried out on 8-core Intel i7 3.6 GHz machines. The whole process - including segmentation, registration with reference images and metric evaluations for a single test image took 7–10 min making it suitable for background processing in large-scale studies and clinical workflows.
Scatter plots of predicted and real scores for 240 segmentations generated with Random Forests of depths 5 (black), 10 (blue) and 40 (red). Metrics are (left to right) Dice similarity coefficient (DSC), mean surface distance (MSD), root-mean-square error (RMS) and Hausdorff distance (HD). Mean absolute error is reported. Correlation coefficients are \(r \approx 0.9, p < 0.001\) for all metrics. Threshold of 0.7 shown on DSC for classification accuracy.
3.1 Automatic Quality Control on UK Biobank Imaging Study
RCA can predict segmentation quality in the absence of ground truth. This makes it attractive for automatic QC in large databases such as those acquired in population imaging. We performed RCA on automatic segmentations of cardiac MR images from the UKBB Imaging StudyFootnote 1 for which manual QC scores have been obtained. These images have a resolution of \(1.83 \times 1.83 \times 10.00\) mm and span \(192 \times 208\) pixels per slice. Thus, they are of lower resolution and have a larger slice thickness than the data in our validation study. The number of slices per scan varies in the range \(\left[ 4 14\right] \) with the majority (89%) having 9–12 slices.
In total, 7,425 cardiac MR images were available to us through the UKBB resource. Each image has been automatically segmented using a probabilistic patch-based multi-atlas approach [13]. As part of a genome-wide association study (GWAS), each automatic segmentation has been checked manually to confirm segmentation quality. The manual QC is based on visual inspection of the basal, mid and apical layers and for each layer a score between 0 and 2 is assigned based on the quality of the class 2 segmentation, the LV myocardium. The total QC score is thus between 0 and 6, where a 6 would be considered as highly accurate segmentation. Where the UKBB images had a poor FOV, the segmentations were immediately discarded for use in the GWAS study: we have given these images a score of \(-1\). We expect that despite the poor FOV of these images, the segmentations itself might still be of good quality. Out of the 7,425 segmented images, 161 have a bad FOV (\(\text {QC}=-1\)) and 43 have an obviously poor segmentation (\(\text {QC}=0\)). There are 2, 13, 50, 307, 2941 and 3908 images having QC scores \([1\ 6]\) respectively. We explored how well RCA based quality predictions correlate with those manual QC scores.
(Top) Comparison of manual, based on LV myocardium, and automatic QC score using RCA on 7,425 images from the UKBB. Class 2 predictions (y-axis) of DSC, MSD, RMS and HD shown (left to right). Correlation is evident between predictions and manual QC scores. (Bottom) A, B, C and D show images with QC score of 1, 2, 5 and 6 respectively. Note that RCA has correctly flagged C as a bad segmentation despite its good (but incorrect) manual QC score.
We performed RCA on all 7,425 segmentations using a reference set of 100 cardiac atlases that were earlier used for the validation experiment. The RCA process took 30–40 min per UKBB image including 100 image registrations. This could be reduced by employing an atlas pre-selection to choose only reference images that are most similar to the test image. Figure 3 shows the relationship between manual QC scores and the predicted DSC, MSD, RMS and HD obtained from RCA. Note, these predictions are for class 2 and not the overall segmentation as this class was the focus of the manual QC. There is an evident correlation between the manual QC score and the RCA predictions. Figure 3 shows a sample of segmentations from the 1, 2, 5 and 6 manual QC scores. With a score of 1, ‘A’ must have an ‘average’ quality segmentation of class 2 (LV myocardium) at either the basal, apical or mid slice while the other layers are clearly poor. Example ‘B’ has highly elongated LV myocardium and cavity leading to high surface-distance metrics and a low DSC. This segmentation has been given a score of 2 because the apical slice is well segmented while the rest is not which is nicely picked up by RCA. In example ‘C’, the segmentation is clearly rotated with respect to the image indicating failed initialisation for the atlas-based approach, but class 2 has a good segmentation in all three regions that gives it its manual QC score of 5. Again, RCA is able to pick up such outliers. For the majority of segmentations with a manual QC of 6, their RCA predictions also indicate high quality. These examples demonstrate RCA’s ability to correctly identify ‘poor’ segmentations when performing assessments over the entire 3D segmentation. It also demonstrates the limitations of manual QC.
4 Conclusion
Reverse classification accuracy had been shown to effectively predict the accuracy of whole-body multi-organ segmentation. We have successfully validated the RCA framework on 3D cardiac MR, demonstrating the robustness of the methodology to different anatomy. RCA has been successful in identifying poor-quality image segmentations with measurements of DSC, MSD, RMS and HD and has shown excellent MAE against all metrics. RCA has also been successful in producing a comparable outcome to a manual quality control procedure on a large database of 7,425 images from the UK Biobank. Predicting segmentation accuracy in the absence of ground truth is a step towards fully automated QC of image analysis pipelines. Future work will investigate the use of RCA in adaptive learning where automatic segmentations with predicted high quality will be used for augmenting training sets. An attractive application is transfer learning and domain adaptation where the automatic, high quality segmentations will help to adjust a learned classifier such as a CNN to a new target domain.
Notes
- 1.
UK Biobank Resource under Application Number 12579.
References
Sudlow, C., Gallacher, J., Allen, N., Beral, V., Burton, P., Danesh, J., Downey, P., Elliott, P., Green, J., Landray, M., Liu, B., Matthews, P., Ong, G., Pell, J., Silman, A., Young, A., Sprosen, T., Peakman, T., Collins, R.: UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12(3), 1–10 (2015)
Shariff, A., Kangas, J., Coelho, L.P., Quinn, S., Murphy, R.F.: Automated image analysis for high-content screening and analysis. J. Biomol. Screen. 15(7), 726–734 (2010)
de Bruijne, M.: Machine learning approaches in medical image analysis: from detection to diagnosis. Med. Image Anal. 33, 94–7 (2016)
Crum, W.R., Camara, O., Hill, D.L.G.: Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imag. 25(11), 1451–1461 (2006)
Taha, A.A., Hanbury, A.: Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med. Imag. 15, 29 (2015)
Valindria, V.V., Lavdas, I., Bai, W., Kamnitsas, K., Aboagye, E.O., Rockall, A.G., Rueckert, D., Glocker, B.: Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE Trans. Med. Imag.PP, 1–1 (2017)
Zhong, E., Fan, W., Yang, Q., Verscheure, O., Ren, J.: Cross validation framework to choose amongst models and datasets for transfer learning. In: Balcázar, J.L., Bonchi, F., Gionis, A., Sebag, M. (eds.) ECML PKDD 2010. LNCS (LNAI), vol. 6323, pp. 547–562. Springer, Heidelberg (2010). doi:10.1007/978-3-642-15939-8_35
Fan, W., Davidson, I.: Reverse testing. In: Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD 2006, p. 147. ACM Press, New York (2006)
Kohlberger, T., Singh, V., Alvino, C., Bahlmann, C., Grady, L.: Evaluating segmentation error without ground truth. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012. LNCS, vol. 7510, pp. 528–536. Springer, Heidelberg (2012). doi:10.1007/978-3-642-33415-3_65
Carapella, V., Jiménez-Ruiz, E., Lukaschuk, E., Aung, N., Fung, K., Paiva, J., Sanghvi, M., Neubauer, S., Petersen, S., Horrocks, I., Piechnik, S.: Towards the semantic enrichment of free-text annotation of image quality assessment for UK biobank cardiac cine MRI scans. In: Carneiro, G., et al. (eds.) LABELS/DLMIA -2016. LNCS, vol. 10008, pp. 238–248. Springer, Cham (2016). doi:10.1007/978-3-319-46976-8_25
Zhang, L., Gooya, A., Dong, B., Hua, R., Petersen, S.E., Medrano-Gracia, P., Frangi, A.F.: Automated quality assessment of cardiac MR images using convolutional neural networks. In: Tsaftaris, S.A., Gooya, A., Frangi, A.F., Prince, J.L. (eds.) SASHIMI 2016. LNCS, vol. 9968, pp. 138–145. Springer, Cham (2016). doi:10.1007/978-3-319-46630-9_14
Zikic, D., Glocker, B., Criminisi, A.: Encoding atlases by randomized classification forests for efficient multi-atlas label propagation. Med. Image Anal. 18(8), 1262–1273 (2014)
Bai, W., Shi, W., O’Regan, D.P., Tong, T., Wang, H., Jamil-Copley, S., Peters, N.S., Rueckert, D.: A probabilistic patch-based label fusion model for multi-atlas segmentation with registration refinement: application to cardiac MR images. IEEE Trans. Med. Imag. 32(7), 1302–1315 (2013)
Acknowledgements
RR is funded by GSK and EPSRC CDT in Medical Imaging (EP/L015226/1); VV by Indonesia Endowment for Education (LPDP) Indonesian Presidential PhD Scholarship; HS by Research Fellowship from Uehara Memorial Foundation; PMM acknowledges support of Imperial Healthcare Trust BRC, EPSRC Centre for Mathematics in Precision Healthcare and MRC.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Robinson, R. et al. (2017). Automatic Quality Control of Cardiac MRI Segmentation in Large-Scale Population Imaging. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10433. Springer, Cham. https://doi.org/10.1007/978-3-319-66182-7_82
Download citation
DOI: https://doi.org/10.1007/978-3-319-66182-7_82
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66181-0
Online ISBN: 978-3-319-66182-7
eBook Packages: Computer ScienceComputer Science (R0)