Abstract
Cochlear implants (CIs) are neural prosthetics that are used to treat sensory-based hearing loss. There are over 320,000 recipients worldwide. After implantation, each CI recipient goes through a sequence of programming sessions where audiologists determine several CI processor settings to attempt to optimize hearing outcomes. However, this process is difficult because there are no objective measures available to indicate what setting changes will lead to better hearing outcomes. It has been shown that a simplified model of electrically induced neural activation patterns within the cochlea can be created using patient CT images, and that audiologists can use this information to determine settings that lead to better hearing performance. A more comprehensive physics-based patient-specific model of neural activation has the potential to lead to even greater improvement in outcomes. In this paper, we propose a method to create such customized electro-anatomical models of the electrically stimulated cochlea. We compare the accuracy of our patient-specific models to the accuracy of generic models. Our results show that the patient-specific models are on average more accurate than the generic models, which motivates the use of a patient-specific modeling approach for cochlear implant patients.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
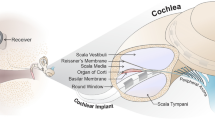
Cochlear implants (CIs) are neural prosthetics that are used to treat sensory- based hearing loss. There are over 320,000 recipients worldwide. CIs have an external and an internal component. The external component is responsible for processing speech and transmitting it to the internal component. The internal component consists of a receiver and an array on which a certain number of electrodes are situated. The electrode array is surgically inserted into the cochlea through a small opening. Sounds are mapped into frequency channels, each corresponding to an electrode. When a sound contains frequencies associated with a channel, the corresponding electrode is activated in order to electrically stimulate the auditory nerve and create the sensation of that sound.
After implantation, each CI recipient goes through a sequence of CI programming sessions to determine CI processor settings, i.e., which electrodes will be activated or deactivated, stimulation levels and frequency bands assigned to each electrode. The programming is one of the factors that plays a significant role on the effectiveness of the CIs. Optimal CI processor settings depend on many factors such as location of electrodes within the cochlea and distance from each electrode to the nerve cells [1, 2]. Thus, it is crucial to correctly localize the electrodes within the cochlea in order to determine the optimal settings that will lead to better hearing outcomes. However, since the electrode array is surgically inserted into the cochlea through a small opening, the intra-cochlear location of the electrodes is usually unknown. In addition, weeks or months of experience with the programmed settings is needed for hearing performance to stabilize enough to be measured reliably. Therefore, the programming process takes several months, and it might not result in optimal settings.
It has been shown by Noble et al. [3] that neural activation caused by CI electrodes can be estimated from CT images by measuring the distance from each electrode to the neural activation sites, and that this information can be used to determine customized CI processor settings. Although this technique has been shown to improve hearing outcomes [2], the indirect estimate of neural activation might be less accurate than a high resolution electro-anatomical model (EAM) of the electrically stimulated cochlea. Three-dimensional EAMs have been used by several different groups in order to investigate the voltage distribution and neural activation within the cochlea [4,5,6,7]. Even though these models have been shown to be useful, they lack the capacity to be applied in vivo, and patient-specific differences cannot be incorporated. It has been previously shown that anatomical shape variations exist [8] and likely lead to different neural activation patterns [9]. For this reason, Malherbe et al. [10] used CT images to construct patient-specific electrical models of CI users. However the model relies on manual point selection as well as approximation of fine scale intra-cochlear structures.
In this study, our aim is to create patient-specific high resolution EAMs using the patient CT image and high resolution resistivity maps constructed from \(\upmu \)CTs of ex vivo cochlea specimens that we can register to the patient CT image. As opposed to a rough approximation of the fine scale structures as is done in [10], we leverage existing segmentation approaches that permit highly accurate localization of structures when creating the model. This is important because it has been shown in [9] that accurate localization is critical to make an accurate model and approximations done at CT resolution are inadequate. We also aim to compare accuracy of generic models, which are currently the community standard, to patient-specific ones.
2 Methods
\(\upmu \)CT images of 9 cadaveric cochlea specimens were acquired using a ScanCo \(\upmu \)CT scanner that produces images with voxel size of 0.036 mm isotropic. Conventional CT images of 5 of the 9 cadaveric cochlea specimens were acquired using a Xoran XCAT scanner with voxel size of 0.3 mm isotropic. The remaining 4 specimens were used in another study which prevented acquisition of conventional images.
The overview of the method proposed in this paper is shown in Fig. 1. In order to make a patient customized model with a new patient image, high resolution ‘resistivity maps’, which are tissue class label maps used to define the electrical resistivity of the tissue in the image, are created from \(\upmu \)CT images of ex-vivo specimens and are projected onto the patient image through a thin-plate spline (TPS) transformation that registers segmentations in the new patient image with segmentations in the \(\upmu \)CTs. A combined resistivity map is created using a majority voting scheme between all of the 9 possible resistivity maps. Using the combined resistivity map and the patient’s known electrode position, a patient-specific model is created. Patient-specific neural activation is then estimated as the current density profile (CDP) along Rosenthal’s Canal (RC) (see Fig. 2), which is where spiral ganglion nerve cells activated by the CI are located. We also create what we refer to as a generic model to compare to our patient-specific one. A generic model is created for a new patient by: (1) mapping the patient electrode positions onto the set of high resolution resistivity maps using a TPS transformation. Each resistivity map is used to estimate a CDP. Then, the generic CDP is computed by averaging CDPs calculated from all 9 models. We create our generic CDP by averaging the results of multiple models, as opposed to using the results from a single model, to avoid biasing the results towards the anatomy of a single individual. In this work, we implemented these proposed models and evaluated their accuracy using a leave-one-out strategy. The following sub-sections detail our approach.
2.1 Creating \(\upmu \)CT Based Electro-Anatomical Model
\(\upmu \)CT images were manually segmented, and 3D meshes of the scala tympani (ST), scala vestibuli (SV) and modiolus (MO) were created as shown in Fig. 3. ST and SV are intra-cochlear cavities filled with perilymph fluid, and MO is where the auditory nerves are located. High resolution resistivity maps were created based on the \(\upmu \)CT images as proposed by Cakir et al. [9]. In brief, a node was defined in the center of each voxel within the field of view of the \(\upmu \)CT image, and a tissue class was assigned to each node depending on their location. The nodes that were enclosed within the ST or SV meshes were assigned to the electrolytic fluid class, and those within the MO mesh were assigned to the neural tissue class. For the remaining nodes, a simple thresholding was applied in order to assign them into either bone, soft tissue or air.
The tissue classes represent different levels of resistivity values where air, bone, soft tissue, neural tissue, and electrolytic fluid have resistivity values of \(\infty \) \(\Omega \)cm, 5000 \(\Omega \)cm, 300 \(\Omega \)cm, 300 \(\Omega \)cm, and 50 \(\Omega \)cm [11], respectively. Finally, for each \(\upmu \)CT image, electrode positions were defined manually at 6 different locations (90, 180, 270, 360, 450, and 540 degree-depths) representative of a typical range of electrode locations in CI recipients. Angular-depth of an electrode is measured as the angle along the spiral of the cochlea, where the round window corresponds to zero degrees (0\(^{\circ }\)) (see Fig. 2). The ground node was placed in the internal auditory canal (IAC) because it is believed that nearly all the current injected via CI electrodes return to the CI ground through the IAC.
2.2 Solving Electro-Anatomical Models
A system of linear equations in the form of \(A\phi = b\) was created to solve Poisson’s equation:
where A is an n \(\times \) n matrix containing the coefficients determined using Eq. 2 and Ohm’s Law. \(\phi \) is an n \(\times \) 1 vector containing the voltage values at each node, and b is an n \(\times \) 1 vector containing the sum of currents entering and leaving each node. The system was solved, using the bi-conjugate gradient method [12].
The tissue in our model was assumed to be purely resistive. Thus, the amount of current that enters a node is equal to the amount of current that leaves the same node, except for the sink and source nodes. Using this assumption and the notation by Whiten [4], the sum of currents entering and leaving the node located at i, j, k can be written as:
which was set equal to \(1\,\upmu \)A if the node was a source, \(-1\,\upmu \)A if it was a sink, and zero for every other node.
2.3 Creating Patient-Specific Electro-Anatomical Models
CT images were automatically segmented using the method developed by Noble et al. [13], which uses an active-shape model based technique. The active shape model is constructed from the ST, SV, and MO surfaces that have been manually defined in the \(\upmu \)CTs, and this allows one-to-one point correspondences to exist between the surfaces manually defined in the \(\upmu \)CT datasets and those automatically localized in the CT datasets. After automatic segmentation, each CT image was manually aligned with its corresponding \(\upmu \)CT image in order for the electrodes defined in the \(\upmu \)CT image space to correspond to the same anatomical location in the CT image space. An example of segmented and aligned \(\upmu \)CT and CT images of a specimen is shown in Fig. 3.
High resolution resistivity maps created using \(\upmu \)CT images were fit to each CT image by leveraging the one-to-one point correspondence property of the active-shape model segmentation. An interpolating TPS-based nonlinear mapping was created using the meshes segmented in the CT and \(\upmu \)CT images as landmarks. TPS defines a non-rigid transformation that minimizes the bending energy between two sets of landmarks [14]. Using a leave-one-out strategy, 8 nonlinear mappings were created between a CT image of one specimen and the \(\upmu \)CT images of the remaining 8 specimens. These nonlinear mappings allowed the construction of 8 different high resolution resistivity maps for each CT image. The tissue class at each pixel in the final map was chosen by majority vote:
where \(z_i\) is the stored candidate tissue class for the \(i^{th}\) resistivity map.
In addition, the nonlinear mapping was used to localize the RC (see Fig. 2) in the newly constructed resistivity map as RC is not visible in CT images due to lack of adequate resolution. Manually segmented RCs in the remaining 8 \(\upmu \)CT images were mapped to the CT image, resulting in 8 different RC segmentations. The final RC segmentation was generated as the average RC of all 8 segmentations. The position of the electrodes for each specimen was determined to be the same position defined in the corresponding registered \(\upmu \)CT for that specimen as described in Sect. 2.1.
2.4 Creating Generic Electro-Anatomical Models
Using a leave-one-out strategy, electrode locations defined in a target specimen image were nonlinearly mapped to the high resolution resistivity maps of the remaining 8 specimens through the corresponding TPS transformations. This produced 8 individual models which were executed resulting in 8 different CDPs. The final generic CDP was determined as the mean across the 8 CDPs. This method was used to create a CDP that is representative of an average cochlea.
2.5 Evaluation
While in-vivo CDP measurement would provide the best ground truth, such measurements are not possible. Thus, we defined the CDPs calculated from the models created using the target specimens’ \(\upmu \)CT images as the ground truth, and compared them to the CDPs calculated using patient-specific and generic models. One potential source of error in creating our models is the accuracy of the automatic anatomy segmentations in the target specimen CT image because the segmentations serve as landmarks for registration with the resistivity maps. To characterize how sensitive our results are to those errors, we also evaluated models constructed using the manual anatomy localizations that we have for the target specimen from its corresponding \(\upmu \)CT image, which provides a baseline for how accurate our models could be given ideal landmark localization.
3 Results
The accuracy of the patient-specific and generic models was quantified as (100% - error), where error is the absolute mean percent difference compared to the ground truth CDP. Table 1 shows the accuracy of the patient-specific and generic models created using manual anatomy localizations for model registration. As shown in the table, patient-specific models are relatively more accurate than generic models, demonstrated by a higher value of accuracy of 87.5% compared to 78.4%, respectively. Table 1 also presents the accuracy of patient-specific and generic models created using automatic landmark localization techniques. On average, patient-specific models are more accurate than generic models, 81.2% compared to 77.2%, respectively.
In addition, the minimum accuracy of the patient-specific model 76.9% is relatively higher than that of the generic model, 66.8%. In general, models created using manual anatomy localizations are more accurate than those created using automatic anatomy localizations. A visual comparison between CDPs calculated from patient-specific, ground truth, and generic models for specimens 4 and 6, the cases where the patient-specific model is the least and the most accurate compared to the generic model, is shown in Fig. 4.
4 Conclusions
To the best of our knowledge, this is the first time that a high resolution patient-specific model was created using CT images and the accuracy of such models was compared to that of generic models. Quantitative and qualitative analysis of the results indicate that improvements in landmark localization could lead to more accurate models and that patient-specific models are on average more accurate than generic models, which is currently the community standard approach. These results motivate the use of patient-specific models and represent a crucial step toward developing and validating the first in vivo patient-specific EAM, which will be used to better customize CI processor settings.
References
Holden, L.K., Finley, C.C., Firszt, J.B., Holden, T.A., Brenner, C., Potts, L.G., Gotter, B.D., Vanderhoof, S.S., Mispagel, K., Heydebrand, G., et al.: Factors affecting open-set word recognition in adults with cochlear implants. Ear Hear. 34(3), 342 (2013)
Noble, J.H., Gifford, R.H., Hedley-Williams, A.J., Dawant, B.M., Labadie, R.F.: Clinical evaluation of an image-guided cochlear implant programming strategy. Audiol. Neurotol. 19(6), 400–411 (2014)
Noble, J.H., Labadie, R.F., Gifford, R.H., Dawant, B.M.: Image-guidance enables new methods for customizing cochlear implant stimulation strategies. IEEE Trans. Neural Syst. Rehabil. Eng. 21(5), 820–829 (2013)
Whiten, D.M.: Electro-anatomical models of the cochlear implant. Ph.D. thesis, Massachusetts Institute of Technology (2007)
Kalkman, R.K., Briaire, J.J., Frijns, J.H.: Current focussing in cochlear implants: an analysis of neural recruitment in a computational model. Hear. Res. 322, 89–98 (2015)
Goldwyn, J.H., Bierer, S.M., Bierer, J.A.: Modeling the electrode-neuron interface of cochlear implants: effects of neural survival, electrode placement, and the partial tripolar configuration. Hear. Res. 268(1), 93–104 (2010)
Hanekom, T.: Three-dimensional spiraling finite element model of the electrically stimulated cochlea. Ear Hear. 22(4), 300–315 (2001)
Avci, E., Nauwelaers, T., Lenarz, T., Hamacher, V., Kral, A.: Variations in microanatomy of the human cochlea. J. Comp. Neurol. 522(14), 3245–3261 (2014)
Cakir, A., Dawant, B.M., Noble, J.H.: Evaluation of a \(\mu \)ct-based electro-anatomical cochlear implant model. In: SPIE Medical Imaging, International Society for Optics and Photonics, pp. 97860M–97860M (2016)
Malherbe, T., Hanekom, T., Hanekom, J.: Constructing a three-dimensional electrical model of a living cochlear implant user’s cochlea. Int. J. Numer. Meth. Biomed. Eng. 32(7), e02751 (2015)
Geddes, L., Baker, L.: The specific resistance of biological material-a compendium of data for the biomedical engineer and physiologist. Med. Biol. Eng. 5(3), 271–293 (1967)
Press, W.H.: Numerical Recipes: The Art of Scientific Computing, 3rd edn. Cambridge University Press, New York (2007)
Noble, J.H., Labadie, R.F., Majdani, O., Dawant, B.M.: Automatic segmentation of intracochlear anatomy in conventional CT. IEEE Trans. Biomed. Eng. 58(9), 2625–2632 (2011)
Bookstein, F.: Thin-plate splines and the decomposition of deformation. IEEE Trans. Pattern Anal. Mach. Intell. 10, 849–865 (1988)
Acknowledgments
This research has been supported by NIH grant R01DC014037. The content is solely the responsibility of the authors and does not necessarily represent the official views of this institute.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Cakir, A., Dawant, B.M., Noble, J.H. (2017). Development of a \(\upmu \)CT-based Patient-Specific Model of the Electrically Stimulated Cochlea. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10433. Springer, Cham. https://doi.org/10.1007/978-3-319-66182-7_88
Download citation
DOI: https://doi.org/10.1007/978-3-319-66182-7_88
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66181-0
Online ISBN: 978-3-319-66182-7
eBook Packages: Computer ScienceComputer Science (R0)