Abstract
Selection of relevant electrodes is of prime importance for developing efficient motor imagery Brain Computer Interface devices. In this paper, we propose a novel spectral clustering based on temporal similarity of electrodes to select a reduced set of relevant electrodes for classification of motor imagery tasks. Further, Stationary common spatial pattern method in conjunction with Composite kernel Support Vector Machine is utilized to develop a decision model. Experimental results demonstrate improvement in classification accuracy in comparison to variants of the common spatial pattern method on publicly available datasets. Friedman statistical test shows that the proposed method significantly outperformed the variants of the common spatial pattern method.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Brain computer interfaces (BCI) enable people with neurological disorders to establish communication and repair lost motor functions by transforming the brain signals into device commands. Non-invasive nature, low measurement cost and high resolution of EEG based BCI has favored its wide use for analysis of brain signals as compared to other modalities. EEG based BCI systems associated with motor imagery has received particular attention that involve visualizing movement of a specific motor part of the body [2]. Motor imagery BCIs use brain dynamics originating in primary sensorimotor area called sensorimotor rhythms (mu and beta rhythms), induced by execution or imagination of hand or leg movement, to translate EEG signals into device commands [2]. During motor imagination or execution, the amplitude of sensorimotor rhythms reduces, which is known as Event-Related Desynchronization (ERD). Increment in the amplitude of sensory-motor rhythms just after the motor imagination or execution is called as Event-Related Synchronization (ERS) [8].
Common spatial pattern (CSP) [2] is a well known spatial filtering method that finds subject specific data dependent spatial filters which maximizes variance of one class and minimizes variance of the other class simultaneously [2]. Extraction of information using the CSP is challenging as this method suffers from the small sample size (SSS) problem [4], i.e. the quantity of electrodes is high and the number of task specific EEG samples is less. In such circumstances, Eigen decomposition of the covariance matrix, whose dimension is number of electrodes x number of electrodes, is highly computational and may lead to imprecision.
To reduce the number of electrodes, the research work [3] has utilized neurological information to segregate the electrodes placed on the brain on the basis of various anatomical areas of brain cortex and select all electrodes of relevant brain areas. However, this method utilizes topographical information based stationary division of electrodes. Further, this approach either uses all electrodes of a chosen cluster or removes all electrodes of a minimum significant cluster. There is a possibility that few of the chosen electrodes may not be pertinent or might be repetitive and few of the removed electrodes of the not chosen brain region might be applicable for recognizing two motor imagery tasks. Hence, in this work, the electrodes are partitioned into many clusters such that each cluster contains a set of similar electrodes to reduce the number of electrodes. For this we use the spectral graph clustering method [7], where the optimal number of clusters is decided using the Davies Bouldin (DB) index criterion [5]. Features are extracted from each cluster using stationary CSP (SCSP) method [6]and composite kernel based support vector machine (CKSVM) is utilised to build a decision model.
The major contributions of this paper include: (i) To select a subject specific reduced subset of relevant electrodes; (ii) To evaluate and compare the performance of the proposed method and variants of CSP on the publicly available datasets; (iii) Friedman test is used to demonstrate that the proposed method significantly outperforms variants of the CSP method. Rest of the article is organized as follows. Section 2 discusses the proposed work. Experimental results are discussed in Sect. 3 and finally, Sect. 4 concludes the article and provides future directions.
2 Proposed Method: Temporal Similarity Based Clustering in Conjunction with CKCSP (TSC-CKCSP)
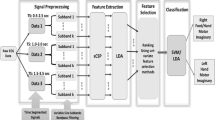
The flow diagram of the proposed model is shown in Fig. 1. A brief description of each step is described as follows:
2.1 Spectral Graph Clustering Based Division of Electrodes
Spectral clustering is a graph theoretic approach to obtain clusters using spectral decomposition of a similarity matrix \(\mathbf S \). To measure similarity between two samples, \(x_i\) and \(x_j\), we have used Gaussian kernel which is defined as:
where \( \sigma \) is a tuning parameter. Let \(\mathbf d \) denotes the vector \(d=[d_1,d_2,\cdots ,.d_N]\) where \(d_i = \sum _{j=1}^N S(i,j)\) measures similarity of \(i^{th}\) sample with all other samples. The larger value of \(d_i\) signifies more similarity of sample \(x_i\) with rest of the samples. Thus, the degree matrix \(\mathbf D \) is given by:
The Laplacian matrix \(\mathbf L \) and normalized Laplacian matrix \(\tilde{\mathbf{L }}\) are defined as:
Eigenvalues and eigenvectors of the normalized Laplacian matrix \(\tilde{\mathbf{L }}\) is solved as:
Let the Eigenvector \(\gamma _2\) correspond to the second smallest Eigenvalue \(\delta _2\). Binary partitioning of electrodes using Eigenvector \(\gamma _2\) is done as follows:
A given cluster is further recursively partitioned using spectral clustering method. To obtain an optimal number of clusters, a well-known Davies Bouldin (DB) criterion is used, which minimizes the ratio of within-cluster dispersion to between-cluster separation. The within-cluster dispersion for \(i^{th}\) cluster, \(Sw_i\) and between-cluster separation for cluster i and cluster j, \(db_{i,j}\) are computed as:
where \(z_i\) is the centroid of the \(i^{th}\) cluster. The similarity measure between cluster i and cluster j and the corresponding DB index are given as:
where \(M_i=\max _{ij}M_{i,j}\) and k is the number of clusters. The number of clusters that provides minimal DB index value is considered optimal. The division of electrodes on both datasets is shown in Fig. 2.
2.2 Stationary Common Spatial Patterns (SCSP)
Relevant features from each cluster of electrodes are computed using SCSP, which is evolved by introducing variations in the Rayleigh criterion function of the traditional CSP technique. Let \(\mathbf {\Sigma _1}\) and \(\mathbf {\Sigma _2}\) be the average covariance matrices of motor imagery left and right hand movement (class 1 and class 2), respectively and \(\mathbf {W}\) is a spatial filter matrix for a given cluster. The Rayleigh criterion maximization function using SCSP is given as:
where \(\mathbf P(W) \) is the introduced penalty term and \(\beta \) is a constant obtained after cross validation method. Further, the features computed from each cluster are transformed to a high dimensional Hilbert kernel space using a Gaussian kernel function to capture the non-linear relations of the extracted features as:
where \({{f}}_{i,l}\) represents features from cluster l for trial i, obtained using SCSP method.
2.3 Feature Selection and Classification
In the proposed method, we have used CKSVM method for classification of motor imagery tasks. CKSVM considers the relevancy of an electrode cluster for recognition of motor imagery tasks.
where \(\alpha _i\) is the classifier parameter, C is a classifier regularization parameter, N is the number of samples and R is the number of clusters. Recursive Feature Elimination (RFE) is used to order the electrode cluster of a specific brain region by calculating the quadratic norm of each cluster of electrodes. Higher is the estimation of the quadratic norm, more important is that cluster to a motor imagery task. The cluster with lowest value of quadratic normal form is removed in each iteration till maximum classification accuracy is achieved.
3 Experimental Setup and Results
The experimental evaluation of the proposed work is performed on publicly available BCI Competition III dataset 4a (Dataset 1) and BCI competition IV dataset Ia (Dataset 2) [3]. Dataset 1 and Dataset 2 consist of motor imagery data acquired from five and seven healthy subjects respectively. The data captured for each trial belongs to the time window of 0.5–2.5 s after the onset of stimulus. Thus, from each electrode, 200 time units are utilized. Whole data is filtered using a [7–30 Hz] bandpass filter. SCSP penalty parameter \(\beta \) = 0.1 was used for all the experiments on both datasets, which is determined using cross-validation. Number of spatial patterns was fixed to r = 1. The average classification accuracy of the proposed method is reported in terms of 10 fold cross-validation run 10 times and is compared to CSP, SCSP and CKSCSP methods as shown in Tables 1 and 2 for subjects of Dataset 1 and Dataset 2, respectively.
We can observe the following from Table 1: (i) The proposed method TSC-CKCSP achieves highest average classification accuracy for Dataset 1; (ii) An overall increment of 7.35%, 4.85%, and 1.1% in classification accuracy as compared to CSP, SCSP and CKSCSP is achieved with our proposed method TSC-CKCSP for Dataset 1. Similarly, the following can be observed from Table 2: (i) The proposed method TSC-CKCSP achieves highest average classification accuracy for Dataset 2 (ii) An overall increment of 12.08%, 3.54%, and 3.17% in classification accuracy as compared to CSP, SCSP and CKSCSP is achieved with our proposed method TSC-CKCSP. The spectral graph clustering is a data centric approach, hence provide relevant and subject specific clusters. Thus, the proposed method performs better than existing methods.
A non-parametric Friedman statistical test [1], is carried out to find the statistical difference between the proposed method and existing methods at significance value of \(\alpha = 0.5\). Table 3 shows the Friedman ranking obtained for each method. P-value computed by Friedman Test is 1.5642 E-6, which signifies that the all methods under comparison are statistical significantly different from each other. Smaller value of Friedman ranking suggests proposed method outperforms variants of CSP.
4 Conclusion
An immense interest has been garnered by Motor imagery BCI due its wide applicability for communication. CSP is a widely used feature extraction technique for motor imagery BCI. However, it suffers from SSS problem due to numerous electrodes and smaller quantity of samples. The proposed method determines a reduced set of relevant electrodes using spectral clustering and CKSVM.
However, the proposed method uses only temporal information of data for division of electrodes and does not consider frequency information for clustering of electrodes. Thus, in future, we will incorporate both spatial, temporal and frequency information for obtaining a reduced and relevant subset of electrodes. Further, RFE in conjunction with SVM utilized in this work is computationally intensive, which requires improvement for real time-applications.
References
Iman, R.L., Davenport, J.M.: Approximations of the critical region of the fbietkan statistic. Commun. Stat.-Theor. Methods 9(6), 571–595 (1980)
Kirar, J.S., Agrawal, R.: Optimal spatio-spectral variable size subbands filter for motor imagery brain computer interface. Proc. Comput. Sci. 84, 14–21 (2016)
Kirar, J.S., Agrawal, R.: Composite kernel support vector machine based performance enhancement of brain computer interface in conjunction with spatial filter. Biomed. Sig. Process. Control 33, 151–160 (2017)
Lu, H., Eng, H.L., Guan, C., Plataniotis, K.N., Venetsanopoulos, A.N.: Regularized common spatial pattern with aggregation for EEG classification in small-sample setting. IEEE Trans. Biomed. Eng. 57(12), 2936–2946 (2010)
Maulik, U., Bandyopadhyay, S.: Performance evaluation of some clustering algorithms and validity indices. IEEE Trans. Patt. Anal. Mach. Intell. 24(12), 1650–1654 (2002)
Samek, W., Vidaurre, C., Müller, K.R., Kawanabe, M.: Stationary common spatial patterns for brain-computer interfacing. J. Neural Eng. 9(2), 026013 (2012)
Von Luxburg, U.: A tutorial on spectral clustering. Stat. Comput. 17(4), 395–416 (2007)
Wolpaw, J., Wolpaw, E.W.: Brain-Computer Interfaces: Principles and Practice. OUP, USA (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Kirar, J.S., Choudhary, A., Agrawal, R.K. (2017). Selection of Relevant Electrodes Based on Temporal Similarity for Classification of Motor Imagery Tasks. In: Shankar, B., Ghosh, K., Mandal, D., Ray, S., Zhang, D., Pal, S. (eds) Pattern Recognition and Machine Intelligence. PReMI 2017. Lecture Notes in Computer Science(), vol 10597. Springer, Cham. https://doi.org/10.1007/978-3-319-69900-4_12
Download citation
DOI: https://doi.org/10.1007/978-3-319-69900-4_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-69899-1
Online ISBN: 978-3-319-69900-4
eBook Packages: Computer ScienceComputer Science (R0)