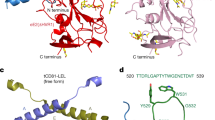
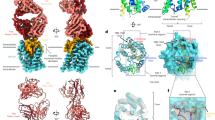
Abstract
Hepatitis C is a global health problem. While many drug companies have active R&D efforts to develop new drugs for treating Hepatitis C virus (HCV), most target the viral enzymes. The HCV glycoprotein E2 has been shown to play an essential role in hepatocyte invasion by binding to CD81 and other cell surface receptors. This paper describes the use of AutoDock to identify ligand binding sites on the large extracellular loop of the open conformation of CD81 and to perform virtual screening runs to identify sets of small molecule ligands predicted to bind to two of these sites. The best sites selected by AutoLigand were located in regions identified by mutational studies to be the site of E2 binding. Thirty-six ligands predicted by AutoDock to bind to these sites were subsequently tested experimentally to determine if they bound to CD81-LEL. Binding assays conducted using surface Plasmon resonance revealed that 26 out of 36 (72 %) of the ligands bound in vitro to the recombinant CD81-LEL protein. Competition experiments performed using dual polarization interferometry showed that one of the ligands predicted to bind to the large cleft between the C and D helices was also effective in blocking E2 binding to CD81-LEL.






Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Ferrari C, Urbani S, Penna A, Cavalli A, Valli A, Lamonaca V, Bertoni R, Boni C, Barbieri K, Uggeri J, Fiaccadori F (1999) Immunopathogenesis of hepatitis C virus infection. J Hepat 31(Supplement 1):3–8
Bartenschlager R (1999) The NS3/4A proteinase of the hepatitis C virus: unraveling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat 6:165–181
Lesburg CA, Radfar R, Weber PC (2000) Recent advances in the analysis of HCV NS5B RNA-dependent RNA polymerase. Curr Opin Investig Drugs 1:289–296
Welbourn S, Pause A (2007) The hepatitis C virus NS2/3 protease. Curr Issues Mol Biol 9:63–69
Venkatraman S, Njoroge FG (2009) Macrocyclic inhibitors of HCV NS3 protease. Expert Opin Ther Pat 19:1277–1303
Enomoto M, Tamori A, Kawada N (2009) Emerging antiviral drugs for hepatitis C virus. Rev Recent Clin Trials 4:179–184
Chary A, Holodniy M (2010) Recent advances in hepatitis C virus treatment: review of HCV protease inhibitor clinical trials. Rev Recent Clin Trials 5:158–173
Sharma SD (2010) Hepatitis C virus: molecular biology and current therapeutic options. Indian J Med Res 131:17–34
Stoll-Keller F, Barth H, Fafi-Kremer S, Zeisel MB, Baumert TF (2009) Development of hepatitis C virus vaccines: challenges and progress. Expert Rev Vaccines 8:333–345
Dubuisson J (2007) Hepatitis C virus proteins. World J Gastroenterol 13:2406–2415
Budkowska A (2009) Mechanism of cell infection with hepatitis C virus (HCV)—a new paradigm in virus-cell interaction. Pol J Microbiol 58:93–98
Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL (2003) Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem 278:41624–41630
Bartosch B, Cosset FL (2006) Cell entry of hepatitis C virus. Virology 348:1–12
Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S (1998) Binding of hepatitis C virus to CD81. Science 282:938–941
Levy S, Todd SC, Maecker HT (1998) CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Ann Rev Immunol 16:89–109
Petracca R, Falugi F, Galli G, Norais N, Rosa D, Campagnoli S, Burgio V, Di Stasio E, Giardina B, Houghton M, Abrignani S, Grandi G (2000) Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol 74:4824–4830
Higginbottom A, Quinn ER, Kuo CC, Flint M, Wilson LH, Bianchi E, Nicosia A, Monk PN, McKeating JA, Levy S (2000) Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol 74:3642–3649
Zhang YY, Zhang BH, Ishii K, Liang TJ (2010) Novel function of CD81 in controlling hepatitis C virus replication. J Virol 84:3396–3407
Drummer HE, Wilson KA, Poumbourios P (2002) Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J Virol 76:11143–11147
VanCompernolle SE, Wiznycia AV, Rush JR, Dhanasekaran M, Baures PW, Todd SC (2003) Small molecule inhibition of hepatitis C virus E2 binding to CD81. Virology 314:371–380
Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M (2001) CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J 20:12–18
Kitadokoro K, Galli G, Petracca R, Falugi F, Grandi G, Bolognesi M (2001) Crystallization and preliminary crystallographic studies on the large extracellular domain of human CD81, a tetraspanin receptor for hepatitis C virus. Acta Crystallogr D Biol Crystallogr 57:156–158
Neugebauer A, Klein CDP, Hartmann RW (2004) Protein-dynamics of the putative HCV receptor CD81 large extracellular loop. Bioorg Med Chem Lett 14:1765–1769
Balhorn R, Hok S, Burke PA, Lightstone FC, Cosman M, Zemla A, Mirick G, Perkins J, Natarajan A, Corzett M, DeNardo SJ, Albrecht H, Gregg JP, DeNardo GL (2007) Selective high-affinity ligand antibody mimics for cancer diagnosis and therapy: initial application to lymphoma/leukemia. Clin Cancer Res 13:5621s–5628s
AutoDock website: http://autodock.scripps.edu
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AK (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semi empirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152
Huey R, Goodsell DS, Morris GM, Olson AJ (2004) Grid-based hydrogen bond potentials with improved directionality. Lett Drug Des Discov 1:178–183
Harris R, Olson AJ, Goodsell DS (2008) Automated prediction of ligand binding sites in proteins. Proteins 70:1506–1517
Morris GM, Huey R, Olson A (2008) Using autodock for ligand-receptor docking. Curr Protoc Bioinforma 8–14
NBCR website: https://www.nbcr.net/pub/wiki/index.php?title=Main_Page
Sanner MF (1999) Python: a programming language for software integration and development. J Mol Graph Model 17:57–61
Holzer M, Ziegler S, Neugebauer A, Kronenberger B, Klein CD, Hartmann RW (2008) Structural modifications of salicylates: inhibitors of human CD81-receptor HCV-E2 interaction. Arch Pharm (Weinheim) 341:478–484
Acknowledgments
This work was supported by The Nadhmi Auchi Fellowship, The American University in Cairo, awarded to R. Al Olaby. Computer time was provided by the National Biomedical Computational Resource at the University of California, San Diego. We would like to thank Dr. Arthur Olson’s molecular graphics laboratory at The Scripps Research Institute for supporting the AutoDock tutorial at Scripps Research Institute and providing assistance with AutoDock 4.2 and AutoDock Tools 1.5.6. We would also like to thank Dr. Shoshana Levy (Stanford University) for generously providing us with the human GST-CD81-LEL protein. Biacore support and instrument use was provided by the Protein Expression Center at Caltech. The ligands tested in this study were provided by the National Cancer Institute through its Developmental Therapeutics Program. Special thanks goes to Dr. David Tirrell (Tirrell lab-Caltech) and Nancy Aitken (Social Entrepreneur) for their great support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Olaby, R.A., Azzazy, H.M., Harris, R. et al. Identification of ligands that target the HCV-E2 binding site on CD81. J Comput Aided Mol Des 27, 337–346 (2013). https://doi.org/10.1007/s10822-013-9649-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-013-9649-3