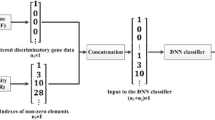
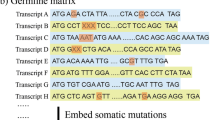
Abstract
It is now well-known that genetic mutations contribute to development of tumors, in which at least 15% of cancer patients experience a causative genetic abnormality including De Novo somatic point mutations. This highlights the importance of identifying responsible mutations and the associated biomarkers (e.g., genes) for early detection in high-risk cancer patients. The next-generation sequencing technologies have provided an excellent opportunity for researchers to study associations between De Novo somatic mutations and cancer progression by identifying cancer subtypes and subtype-specific biomarkers. Simple linear classification models have been used for somatic point mutation-based cancer classification (SMCC); however, because of cancer genetic heterogeneity (ranging from 50 to 80%), high data sparsity, and the small number of cancer samples, the simple linear classifiers resulted in poor cancer subtypes classification. In this study, we have evaluated three advanced deep neural network-based classifiers to find and optimized the best model for cancer subtyping. To address the above-mentioned complexity, we have used pre-processing clustered gene filtering (CGF) and indexed sparsity reduction (ISR), regularization methods, a Global-Max-Pooling layer, and an embedding layer. We have evaluated and optimized the three deep learning models CNN, LSTM, and a hybrid model of CNN + LSTM on publicly available TCGA-DeepGene dataset, a re-formulated subset of The Cancer Genome Atlas (TCGA) dataset and tested the performance measurement of these models is 10-cross-validation accuracy. Evaluating all the three models using a same criterion on the test dataset revealed that the CNN, LSTM, and CNN + LSTM have 66.45% accuracy, 40.89% accuracy, and 41.20% accuracy in somatic point mutation-based cancer classification. Based on our results, we propose the CNN model for further experiments on cancer subtyping based on DNA mutations.













Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Data availability
The dataset analyzed during the current study are available in the DeepGene repository, https://github.com/yuanyc06/deepgene.
References
Aarøe J, Lindahl T, Dumeaux V, Sæbø S, Tobin D, Hagen N, Skaane P, Lönneborg A, Sharma P, Børresen-Dale A-L (2010) Gene expression profiling of peripheral blood cells for early detection of breast cancer. Breast Cancer Res 12:1–11
Abdel-Hamid O, Mohamed A-R, Jiang H, Deng Li, Penn G, Dong Yu (2014) Convolutional neural networks for speech recognition. IEEE/ACM Trans on Audio, Speech, Lang Process 22:1533–1545
Alinejad-Rokny H, Anwar F, Waters SA, Davenport MP, Ebrahimi D (2016) Source of CpG depletion in the HIV-1 genome. Mol Biol Evol 33:3205–3212
Alinejad-Rokny H, Ghavami R, Rabiee HR, Rezaei N, Tam KT, Forrest AR (2020) MaxHiC: robust estimation of chromatin interaction frequency in Hi-C and capture Hi-C experiments. bioRxiv 2020(8):15454
Asrol M, Papilo P, Gunawan FE (2021) Support vector machine with K-fold validation to improve the industry’s sustainability performance classification. Procedia Computer Sci 179:854–862
Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A (2008) Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116:597–602
Bayati M, Rabiee HR, Mehrbod M, Vafaee F, Ebrahimi D, Forrest AR, Alinejad-Rokny H (2020) CANCERSIGN: a user-friendly and robust tool for identification and classification of mutational signatures and patterns in cancer genomes. Sci Rep 10:1–11
Browne RP, McNicholas PD, Sparling MD (2011) Model-based learning using a mixture of mixtures of Gaussian and uniform distributions. IEEE Trans Pattern Anal Mach Intell 34:814–817
Cai Z, Lizhe X, Yi S, Mohammad RS, Randy G, Guohui L. (2006) Using gene clustering to identify discriminatory genes with higher classification accuracy. In Sixth IEEE Symposium on BioInform and BioEng (BIBE'06), 235–42. IEEE
Chanu MM, Thongam K (2021) Computer-aided detection of brain tumor from magnetic resonance images using deep learning network. J Ambient Intell Humaniz Comput 12:6911–6922
Cheng J-Z, Ni D, Chou Y-H, Qin J, Tiu C-M, Chang Y-C, Huang C-S, Shen D, Chen C-M (2016) Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep 6:1–13
Cho J-H, Lee D, Park JH, Lee I-B (2003) New gene selection method for classification of cancer subtypes considering within-class variation. FEBS Lett 551:3–7
Chow, Chi K, Hailong Z, Jessica L, Mark WL, Winston PK, Keith C. (2009) A cooperative feature gene extraction algorithm that combines classification and clustering. In 2009 IEEE Int Conf on Bioinform and Biomed Workshop, 197–202. IEEE
Ciregan D, Ueli M, Jürgen S. (2012) Multi-column deep neural networks for image classification. In 2012 IEEE Conference on Computer Vision and Pattern Recognition, 3642–49. IEEE
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A (2018) Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 24:1559–1567
Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih NNC, Tomaszewski J, González FA, Madabhushi A (2017) Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci Rep 7:1–14
Dashti H, Dehzangi A, Bayati M, Breen J, Lovell N, Ebrahimi D, Alinejad-Rokny H. (2020) Integrative analysis of mutated genes and mutational processes reveals seven colorectal cancer subtypes. bioRxiv
Deepak S, Ameer PM (2021) Automated categorization of brain tumor from mri using cnn features and svm. J Ambient Intell Humaniz Comput 12:8357–8369
Donahue J, Yangqing J, Oriol V, Judy H, Ning Z, Eric T, Trevor D. (2014) Decaf: a deep convolutional activation feature for generic visual recognition. In Int Conf on Mach Learn, 647–55. PMLR
Ebrahimi D, Alinejad-Rokny H, Davenport MP (2014) Insights into the motif preference of APOBEC3 enzymes. PLoS One 9:e87679
Edara DC, Lakshmi PV, Venkatramaphanikumar S, Venkata KKK (2019) Sentiment analysis and text categorization of cancer medical records with LSTM. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-019-01399-8
Fateh A, Fateh M, Abolghasemi V (2021) Multilingual handwritten numeral recognition using a robust deep network joint with transfer learning. Inf Sci 581:479–494
Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F. (2020) Global cancer observatory: cancer today. Lyon: Int Agency Res Cancer; 2018
Gers FA, Schmidhuber J, Cummins F (2000) Learning to forget: continual prediction with LSTM. Neural Comput 12:2451–2471
Ghareyazi A, Mohseni A, Dashti H, Beheshti A, Dehzangi A, Rabiee HR, Alinejad-Rokny H (2021) Whole-genome analysis of de novo somatic point mutations reveals novel mutational biomarkers in pancreatic cancer. Cancers 13:4376
Gong L, Wang C, Li Xi, Chen H, Zhou X (2018) MALOC: a fully pipelined FPGA accelerator for convolutional neural networks with all layers mapped on chip. IEEE Trans Comput Aided Des Integr Circuits Syst 37:2601–2612
Gooneratne SL, Alinejad-Rokny H, Ebrahimi D, Bohn PS, Wiseman RW, O’Connor DH, Kent SJ (2014) Linking pig-tailed macaque major histocompatibility complex class I haplotypes and cytotoxic T lymphocyte escape mutations in simian immunodeficiency virus infection. J Virol 88:14310–14325
Habibi M, Weber L, Neves M, Wiegandt DL, Leser U (2017) Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 33:i37–i48
He K, Xiangyu Z, Shaoqing R, Jian S. (2016) Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770–78
Heidari R, Akbariqomi M, Asgari Y, Ebrahimi D, Alinejad-Rokny H (2021) A systematic review of long non-coding RNAs with a potential role in breast cancer. Mutat Res/Rev Mutat Res 787:108375
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18:1527–1554
Hinton GE, Nitish S, Alex K, Ilya S, Ruslan RS. (2012) Improving neural networks by preventing co-adaptation of feature detectors, arXiv preprint arXiv:1207.0580
Hosseinpoor M, Parvin H, Nejatian S, Rezaie V, Bagherifard K, Dehzangi A, Alinejad-Rokny H (2020) Proposing a novel community detection approach to identify cointeracting genomic regions. Math Biosci Eng 17:2193–2217
Huang J, Vivek R, Chen S, Menglong Z, Anoop K, Alireza F, Ian F, Zbigniew W, Yang S, Sergio G. (2017) Speed/accuracy trade-offs for modern convolutional object detectors. In Proceedings of the IEEE conference on computer vision and pattern recognition, 7310–11
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Xiang Du (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127:118–126
Inan O, Uzer MS (2021) A method of classification performance improvement via a strategy of clustering-based data elimination integrated with k-Fold cross-validation. Arab J Sci Eng 46:1199–1212
Jalali Y, Fateh M, Rezvani M, Abolghasemi V, Anisi MH (2021) ResBCDU-Net: a deep learning framework for lung CT image segmentation. Sensors 21:268
Javanmard R, JeddiSaravi K, Alinejad-Rokny H (2013) Proposed a new method for rules extraction using artificial neural network and artificial immune system in cancer diagnosis. J Bionanosci 7:665–672
Jia AD, Zhengyi Li B, Chuanwang C, Zhang. (2020) Detection of cervical cancer cells based on strong feature CNN-SVM network. Neurocomputing 411:112–127
Kalantari A, Kamsin A, Shamshirband S, Gani A, Alinejad-Rokny H, Anthony T (2018) Computational intelligence approaches for classification of medical data: state-of-the-art, future challenges and research directions. Neurocomputing 276:2–22 (%J Neurocomputing Chronopoulos)
Khan SU, Islam N, Jan Z, Din IU, Rodrigues JJPC (2019) A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett 125:1–6
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
Kurman RJ, Kala V, Richard R, Wu TC, Ie-Ming S (2008) Early detection and treatment of ovarian cancer: shifting from early stage to minimal volume of disease based on a new model of carcinogenesis. Am J Obstetrics Gyneco 198:351–56
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86:2278–2324
Liang G, Hong H, Xie W, Zheng L (2018) Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access 6:36188–36197
Lin M, Qiang C, Shuicheng Y. (2013) Network in network, arXiv preprint arXiv:1312.4400
Liu W, Wang Z, Liu X, Zeng N, Liu Y, Alsaadi FE (2017) A survey of deep neural network architectures and their applications. Neurocomputing 234:11–26
Nguyen LD, Ruihan G, Dongyun L, Zhiping L (2019) Biomedical image classification based on a feature concatenation and ensemble of deep CNNs. J Ambient Intell Human Comput 10:1–13. https://doi.org/10.1007/s12652-019-01276-4
Niu H, Khozouie N, Parvin H, Alinejad-Rokny H, Beheshti A, Mahmoudi MR (2020) An ensemble of locally reliable cluster solutions. Appl Sci 10:1891
Parvin H, Alinejad-Rokny H, Minaei-Bidgoli B (2011a) Detection of cancer patients using an innovative method for learning at imbalanced datasets. International conference on rough sets and knowledge technology. Springer, Berlin Heidelberg, pp 376–381
Parvin H, Minaei B, Alizadeh H, Beigi A (2011b) A novel classifier ensemble method based on class weightening in huge dataset. In international symposium on neural networks. Springer, Heidelberg, pp 144–150
Parvin H, MirnabiBaboli M, Alinejad-Rokny H (2015) Proposing a classifier ensemble framework based on classifier selection and decision tree. Eng Appl Artif Intell 37:34–42
Qaiser T, Tsang Y-W, Epstein D, Rajpoot N (2017) Tumor segmentation in whole slide images using persistent homology and deep convolutional features. Annual conference on medical image understanding and analysis. Springer, Heidelberg, pp 320–329
Rajaei P, Jahanian KH, Beheshti A, Band SS, Dehzangi A, Alinejad-Rokny H (2021) VIRMOTIF: A user-friendly tool for viral sequence analysis. Genes 12:186
Renith G, Senthilselvi A (2020) Accuracy improvement in diabetic retinopathy detection using DLIA. J Adv Res Dyn Control Syst 12(4):133–149. https://doi.org/10.5373/JARDCS/V12I4/20201426
Sankareswaran SP, Krishnan M (2022) Unsupervised end-to-end brain tumor magnetic resonance image registration using RBCNN: rigid transformation, B-spline transformation and convolutional neural network. Curr Med Imaging 18(4):387–397
Shamshirband S, Mahdis F, Abdollah D, Anthony TC, Hamid A-R (2021) A review on deep learning approaches in healthcare systems: taxonomies, challenges, and open issues. J Biomed Inform 113:103627
Sharif RA, Hossein A, Josephine S, Stefan C. (2014) CNN features off-the-shelf: an astounding baseline for recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops, 806–13
Sharifrazi D, Alizadehsani R, Joloudari JH, Shamshirband S, Hussain S, Sani ZA, Alinejad-Rokny H. (2020) CNN-KCL: automatic myocarditis diagnosis using convolutional neural network combined with k-means clustering, preprints, 2020
Shaukat F, Raja G, Ashraf R, Khalid S, Ahmad M, Ali A (2019) Artificial neural network based classification of lung nodules in CT images using intensity, shape and texture features. J Ambient Intell Humaniz Comput 10:4135–4149
Shen D, Guorong Wu, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248
Shen D, Guoyin W, Wenlin W, Martin RM, Qinliang S, Yizhe Z, Chunyuan L, Ricardo H, Lawrence C. (2018) Baseline needs more love: on simple word-embedding-based models and associated pooling mechanisms, arXiv preprint arXiv:1805.09843
Simonyan K, Andrew Z. (2014) Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556
Sujitha R, Seenivasagam V (2021) Classification of lung cancer stages with machine learning over big data healthcare framework. J Ambient Intell Humaniz Comput 12:5639–5649
Sun Yi (2015) Deep learning face representation by joint identification-verification. The Chinese University of Hong Kong, Hong Kong
Surya V, Senthilselvi A (2020) A qualitative analysis of the machine learning methods in food adultery: a focus on Milk adulteration detection. J Adv Res Dyn Control Syst 12(7):543–551. https://doi.org/10.5373/JARDCS/V12I7/20202037
Svensén M, Christopher MB (2007) Pattern recognition and machine learning. Springer, Berlin/Heidelberg, Germany
Szegedy C, Wei L, Yangqing J, Pierre S, Scott R, Dragomir A, Dumitru E, Vincent V, Andrew R. (2015) Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1–9
Szegedy C, Vincent V, Sergey I, Jon S, Zbigniew W. (2016) Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2818–26
Thaha MM, Pradeep Mohan Kumar K, Murugan BS, Dhanasekeran S, Vijayakarthick P, Senthil A, Selvi. (2019) Brain tumor segmentation using convolutional neural networks in MRI images. J Med Syst 43:1–10
Tomczak K, Czerwińska P, Wiznerowicz M (2015) The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol 19:A68
Varsamopoulos S, Bertels K, Almudever CG (2019) Comparing neural network based decoders for the surface code. IEEE Trans Comput 69:300–311
Wang J, Lin J, Wang Z (2017) Efficient hardware architectures for deep convolutional neural network. IEEE Trans Circuits Syst I Regul Pap 65:1941–1953
Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril M-F, Ortiz PL, Romero TR, Balacescu O (2006) Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst 98:472–482
Xu J, Luo X, Wang G, Gilmore H, Madabhushi A (2016) A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 191:214–223
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights Imaging 9:611–629
Yang Z, Ran L, Zhang S, Xia Y, Zhang Y (2019) EMS-net: ensemble of multiscale convolutional neural networks for classification of breast cancer histology images. Neurocomputing 366:46–53
Yuan Y, Shi Yi, Li C, Kim J, Cai W, Han Z, Feng DD (2016) DeepGene: an advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinform 17:243–256
Zhu W, Chaochun L, Wei F, Xiaohui X. (2018) Deeplung: deep 3d dual path nets for automated pulmonary nodule detection and classification. In 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), 673–81. IEEE
Acknowledgements
HAR has been supported by UNSW Scientia Program Fellowship. Analysis was made possible with computational resources provided by the BioMedical Machine Learning high performance computing Server with funding from the Australian Government and the UNSW SYDNEY.
Author information
Authors and Affiliations
Contributions
HAR and PP designed the study; PP, MF, MR designed the models. PP, HAR, MF wrote the paper. HAR, MF, MR, and PP edited the manuscript. PP carried out all the analyses, including the statistical analyses, model developments, comparision, etc. PP generated all figures and all tables. All authors have read and approved the final version of the paper.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Parhami, P., Fateh, M., Rezvani, M. et al. A comparison of deep neural network models for cluster cancer patients through somatic point mutations. J Ambient Intell Human Comput 14, 10883–10898 (2023). https://doi.org/10.1007/s12652-022-04351-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-022-04351-5