Abstract
Background
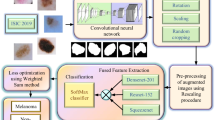
Skin diseases are common health complications around the world. One of the most unsafe types of skin cancer is melanoma. Detection of skin cancer in the early stage can reduce the mortality rate. Manually detecting skin cancer from dermoscopy images is time-consuming, difficult, expensive, and requires experience. Therefore computer-based systems are necessary to make an early finding of skin cancer to plan timely treatment for patients to increase their survival rates. In this work, we introduce a Gaussian filter for noise removal. Furthermore, we applied the Grabcut segmentation technique to segment the affected lesions. Then we applied the segmented image into Convolutional Neural Network (CNN) with three hidden layers, including a flat and dense layer. The proposed CNN model uses the Adam optimizer to deliver the best performance with an accuracy value of 97.50%. In this study, we used 3500 images containing 500 images of each class. The data is further split into training and validation data as 75% and 25%. Based on the proposed model described in Fig. 2, segmented images are of size (32,32,3) as an input to the CNN model, which contains three hidden layers. The idea is convoluted, using 3 × 3 filters of 256,1n28 and 64, respectively, on each hidden layer. This Paper used the Convolutional Neural Network based method proposed for seven types of melanoma classification. It aids Skin cancer classifications that should be detectable by both patients and clinicians. By the experimental and evaluation, it can be said the designed model can be considered as a benchmark for skin cancer classification.








Similar content being viewed by others
Data availability
Dataset will be made available on request.
References
Cancer Research UK. 2012. Cancer World Wide - the global picture. Retrieved March 16, 2019 from https://www.cancerresearchuk.org/health-professional/cancer-statistics/worldwide-cancer. Accessed 5 Jan 2023.
World Health Organization. 2019. Skin Cancer. Retrieved March 16, 2019 from http://www.who.int/en/.
Dong Y, Wang L, Cheng S, Li Y. FAC-Net: feedback attention network based on context encoder network for skin lesion segmentation. Sensors. 2021;21:5172.
Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the us population, 2012. JAMA Dermatol. 2015;151:1081–6.
Stern RS. Prevalence of a history of skin cancer in 2007: results of an incidence-based model. Arch Dermatol. 2010;146:279–82.
Codella NC, Nguyen Q-B, Pankanti S, Gutman DA, Helba B, Halpern AC, Smith J. R Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev. 2017;61:5:1-5:15.
Naik PP. Cutaneous malignant melanoma: a review of early diagnosis and management. World J Oncol. 2021;12:7.
Alhaisoni M, Tariq U, Hussain N, Majid A, Damaševiˇcius R, Maskeliūnas R. COVID-19 case recognition from chest CT images by deep learning, entropy-controlled firefly optimization, and parallel feature fusion. Sensors. 2021;21:7286.
Khan MA, Muhammad K, Sharif M, Akram T, Kadry S. Intelligent fusion-assisted skin lesion localization and classification for smart healthcare. Neural Comput Appl. 2021;36:1–16.
Adegun A, Viriri S. Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art. Artif Intell Rev. 2020;54:811–41.
Ren Y, Yu L, Tian S, Cheng J, Guo Z, Zhang Y. Serial attention network for skin lesion segmentation. J Ambient Intell Humaniz Comput. 2021;13:1–12.
Yu Z, Jiang X, Zhou F, Qin J, Ni D, Chen S, Lei B, Wang T. Melanoma recognition in dermoscopy images via aggregated deep convolutional features. IEEE Trans Biomed Eng. 2018;66:1006–16.
Aziz S, Bilal M, Khan MU, Amjad F. Deep learning-based automatic morphological classification of leukocytes using blood smears. In: Proceedings of the 2020 International Conference on Electrical, Communication, and Computer Engineering (ICECCE), Istanbul, Turkey, 14–15 April 2020; pp. 1–5.
Khan MA, Zhang Y-D, Sharif M, Akram T. Pixels to classes: intelligent learning framework for multiclass skin lesion localization and classification. Comput Electr Eng. 2021;90: 106956.
Navarro F, Escudero-Vinolo M, Bescos J. Accurate segmentation and registration of skin lesion images to evaluate lesion change. IEEE J Biomed Health Inform. 2018;23:501–8.
Hosny KM., Kassem MA, Foaud MM. Skin cancer classification using deep learning and transfer learning. In: Proceedings of the 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), Cairo, Egypt, 20–22 December 2018; pp. 90–93.
Vidya M, Karki MV. Skin cancer detection using machine learning techniques. In: 2020 IEEE International Conference on electronics, computing and communication technologies (CONECCT). IEEE, 2020; pp. 1–5.
Pham TC, Tran GS, Nghiem TP, Doucet A, Luong CM, Hoang V-D. A comparative study for classification of skin cancer. In: 2019 International Conference on System Science and Engineering (ICSSE). IEEE, 2019; pp. 267–272
Mhaske H, Phalke D. Melanoma skin cancer detection and classification based on supervised and unsupervised learning. In: 2013 International conference on Circuits, Controls and Communications (CCUBE). IEEE, 2013; pp. 1–5.
Garnavi R, Aldeen M, Celebi ME, Varigos G, Finch S. Border detection in dermoscopy images using hybrid thresholding on optimized color channels. Comput Med Imaging Graph. 2011;35(2):105–15.
Hoang L, Lee SH, Lee EJ, Kwon KR. Multiclass skin lesion classification using a novel lightweight deep learning framework for Smart healthcare. Appl Sci. 2022;12(5):2677.
Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018;5: 180161.
A. Swain, Noise filtering in digital image processing, medium, 3, 2018, [Online]. Available: https://medium.com/image-vision/noise-filtering-in-digitalimage-processing-d12b5266847c. Accessed 20 Dec 2020.
Thurnhofer-Hemsi K, López-Rubio E, Domínguez E, Elizondo DA. Skin lesion classification by ensembles of deep convolutional networks and regularly spaced shifting. IEEE Access. 2021;9:112193–205.
Chaturvedi SS, Tembhurne JV, Diwan T. A multi-class skin cancer classification using deep convolutional neural networks. Multimed Tools Appl. 2020;79(39):28477–98.
Srinivasu PN, et al. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors. 2021;21(8):2852.
Sindhwani N, Yadav V, et al. Performance analysis of deep neural networks using computer vision. EAI Endorsed Trans Ind Netw Intell Syst. 2021;8(29e3):1–11. https://doi.org/10.4108/eai.13-10-2021.171318.
Deshwal V, Yadav V, et al. Estimating COVID-19 cases using machine learning regression algorithms. Recent Adv Electr Electron Eng. 2022;15(5):390–400. https://doi.org/10.2174/2352096515666220610155214.
Rahul M, Yadav V, et al. A new hybrid approach for efficient emotion recognition using deep learning. Int J Electr Electron Res. 2022;10(1):18–22. https://doi.org/10.37391/IJEER.100103.
Kumar A, Sarkar S, Pradhan C. Malaria disease detection using CNN technique with SGD, RMSprop and ADAM Optimizers. In: Dash S, Acharya B, Mittal M, Abraham A, Kelemen A, editors. Deep learning techniques for biomedical and health informatics. Studies in BIG DATA, vol. 68. Cham: Springer; 2020. https://doi.org/10.1007/978-3-030-33966-1_11.
Hassan E, Shams MY, Hikal NA, et al. The effect of choosing optimizer algorithms to improve computer vision tasks: a comparative study. Multimed Tools Appl. 2023;82:16591–633. https://doi.org/10.1007/s11042-022-13820-0.
Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1-score and accuracy in binary classification evaluation. ChiccoJurman BMC Genom. 2020;21:1–13.
Author information
Authors and Affiliations
Contributions
SV: Conceptualization, methodology, and data curation; MK: formal analysis and writing. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Verma, S., Kumar, M. Automatic Classification of Melanoma Using Grab-Cut Segmentation & Convolutional Neural Network. SN COMPUT. SCI. 5, 591 (2024). https://doi.org/10.1007/s42979-024-02949-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02949-4