Abstract
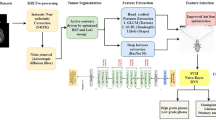
Neuroimaging plays an elemental role in disease detection in the domain of medical science. Brain magnetic resonance imaging (MRI) helps to detect chronic diseases such as brain tumors, strokes, and dementia. It is a nonintrusive and sensitive method for evaluating brain tumors. Numerous deep learning techniques have been proposed to analyze brain tumors as they revolutionize feature selection by automatically extracting relevant features, outperforming traditional methods in accuracy and speed. Our paper proposes a first-of-its-kind, two-stage framework for classifying brain tumors from structural MRI scans. In the first stage, a pre-trained convolutional neural network has been used to extract relevant features, considerably reducing training time and the need for extensive hardware. Next, a filter-based deep feature selection method narrows down the high-dimensional features obtained from the previous stage, minimizing computational load and overfitting risks. Finally, a polynomial-kernel Support vector machine performs multi-class classification. We have also employed fivefold cross-validation to ensure reliable results that are not overly sensitive to specific training or testing data. On the first dataset, this paper achieved 98.17% classification accuracy, 97.92% precision, 97.95% recall, and an F1-score of 97.92% while simultaneously reducing 25% of the extracted features. The approach has also been tested on two additional brain tumor datasets, giving classification accuracies of 99.46% and 98.70%. These promising results underscore the potential of our lightweight framework’s robust nature and generalization capabilities, making it suitable for deployment in real-time environments with limited technological resources.





















Similar content being viewed by others
Data availability
The benchmarked datasets used and analyzed in this study are publicly available at: Brain Tumor MRI Dataset [20]: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset. Crystal Clean: Brain Tumors MRI Dataset [21]: https://www.kaggle.com/datasets/mohammadhossein77/brain-tumors-dataset. Figshare: Brain Tumors MRI Dataset [22]: https://www.kaggle.com/datasets/denizkavi1/brain-tumor.
Source code availability
The source codes related to the current work are made publicly available: https://github.com/Utathyaworks/Brain_Tumor_Classification.
References
Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. https://doi.org/10.1007/s00401-016-1545-1.
Kang J, Ullah Z, Gwak J. MRI-based brain tumor classification using ensemble of deep features and machine learning classifiers. Sensors. 2021;21(6):2222. https://doi.org/10.3390/s21062222.
Gao H, Jiang X. Progress on the diagnosis and evaluation of brain tumors. Cancer Imaging. 2013;13(4):466–481. https://doi.org/10.1102/1470-7330.2013.0039.
Alzubaidi L, Santamar'ia J, Manoufali M, Mohammed BJ, Fadhel MA, Zhang J, Al-timemy AH, Al-Shamma O, Duan Y. MedNet: pre-trained convolutional neural network model for the medical imaging tasks. arXiv: abs/2110.06512. 2021.
Saranya C, Janakiraman GP, Jayalakshmi P, Pavithra E. Brain tumor identification using deep learning. Mater Today Proc. 2021. https://doi.org/10.1016/j.matpr.2020.11.555.
Nandurbarkar S, Singh A, Bute HKN, GiteRajamani SK. Binary classification of brain tumor using early and late fusion. Int J Intell Syst Appl Eng. 2024;12(10s):402–14.
Gómez-Guzmán MA, Jiménez-Beristaín L, García-Guerrero EE, López-Bonilla OR, Tamayo-Perez UJ, Esqueda-Elizondo JJ, Palomino-Vizcaino K, Inzunza-González E. Classifying brain tumors on magnetic resonance imaging by using convolutional neural networks. Electronics. 2023;12(4):955. https://doi.org/10.3390/electronics12040955.
Chattopadhyay S, Singh PK, Ijaz MF, et al. SnapEnsemFS: a snapshot ensembling-based deep feature selection model for colorectal cancer histological analysis. Sci Rep. 2023;13:9937. https://doi.org/10.1038/s41598-023-36921-8.
Marik A, Chattopadhyay S, Singh P. A hybrid deep feature selection framework for emotion recognition from human speeches. Multim Tools Appl. 2022. https://doi.org/10.1007/s11042-022-14052-y.
Sheikh KH, Ahmed S, Mukhopadhyay K, Singh PK, Yoon JH, Geem ZW, Sarkar R. EHHM: Electrical harmony based hybrid meta-heuristic for feature selection. IEEE Access. 2020;8:158125–41. https://doi.org/10.1109/ACCESS.2020.3019809.
Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Adv Neural Inf Process Syst. 2014;27.
Bhattacharya D, Sharma D, Kim W, Ijaz MF, Singh PK. Ensem-HAR: an ensemble deep learning model for smartphone sensor-based human activity recognition for measurement of elderly health monitoring. Biosensors. 2022;12(6):393. https://doi.org/10.3390/bios12060393.
Das SG, Singh PK, Ahmadian A, Senu N, Sarkar R. A hybrid meta-heuristic feature selection method for identification of Indian spoken languages from audio signals. IEEE Access 2020;8:181432–49. https://doi.org/10.1109/ACCESS.2020.3028241.
Das S, Singh PK, Bhowmik S, Sarkar R, Nasipuri M. A harmony search based wrapper feature selection method for holistic bangla word recognition. Procedia Comput Sci. 2016;89:395–403. https://doi.org/10.1016/j.procs.2016.06.087(ISSN 1877-0509).
Yu L, Liu H. Feature selection for high-dimensional data: a fast correlation-based filter solution. In: Fawcett T, Mishra N, editors. In: Proceedings, twentieth international conference on machine learning, vol. 2. 2003. pp. 856–63.
Rajeshwari J, Sughasiny M. Modified filter based feature selection technique for dermatology dataset using beetle swarm optimization. EAI Endorsed Trans Scal Inf Syst. 2022;10(2): e1. https://doi.org/10.4108/eetsis.vi.1998.
Sahoo KK, Ghosh R, Mallik S, et al. Wrapper-based deep feature optimization for activity recognition in the wearable sensor networks of healthcare systems. Sci Rep. 2023;13:965. https://doi.org/10.1038/s41598-022-27192-w.
Ghosh KK, Singh PK, Hong J, Geem ZW, Sarkar R. Binary social mimic optimization algorithm with X-shaped transfer function for feature selection. IEEE Access. 2020;8:97890–906. https://doi.org/10.1109/ACCESS.2020.2996611.
Tan M, Le Q. Efficientnet: rethinking model scaling for convolutional neural networks. In: International conference on machine learning. PMLR. 2019. pp. 6105–14.
Nickparvar M. Brain Tumor MRI Dataset. Kaggle. 2021. https://doi.org/10.34740/KAGGLE/DSV/2645886.
Hashemi SMH. Crystal Clean: Brain Tumors MRI Dataset. Kaggle. 2023. https://doi.org/10.34740/KAGGLE/DS/3505991.
Kavi D. Brain tumor image dataset. Kaggle. 2021. https://www.kaggle.com/datasets/denizkavi1/brain-tumor.
Ge C, Gu IY-H, Jakola AS, Yang J. Enlarged training dataset by pairwise GANs for molecular-based brain tumor classification. IEEE Access. 2020;8:22560–70. https://doi.org/10.1109/ACCESS.2020.2969805.
Yeafi A, Islam M, Mondal S, Ishraq K, Nashad KM, Salah Md, Yusuf U. A semi-supervised approach for brain tumor classification using Wasserstein generative adversarial network with gradient penalty. https://doi.org/10.1109/EICT61409.2023.10427898.
Saeedi S, Rezayi S, Keshavarz H, et al. MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med Inform Decis Mak. 2023;23:16. https://doi.org/10.1186/s12911-023-02114-6.
Raza A, Ayub H, Khan JA, Ahmad I, S. Salama A, Daradkeh YI, Javeed D, Ur Rehman A, Hamam H. A hybrid deep learning-based approach for brain tumor classification. Electronics. 2022;11(7):1146. https://doi.org/10.3390/electronics11071146.
Ali MU, Hussain SJ, Zafar A, Bhutta MR, Lee SW. WBM-DLNets: wrapper-based metaheuristic deep learning networks feature optimization for enhancing brain tumor detection. Bioengineering (Basel). 2023;10(4):475. https://doi.org/10.3390/bioengineering10040475. (PMID: 37106662; PMCID: PMC10135892).
Rasheed Z, Ma Y-K, Ullah I, Ghadi YY, Khan MZ, Khan MA, Abdusalomov A, Alqahtani F, Shehata AM. Brain tumor classification from MRI using image enhancement and convolutional neural network techniques. Brain Sci. 2023;13:1320. https://doi.org/10.3390/brainsci13091320.
Ibrahim R, Ghnemat R, Abu A-H. Improving Alzheimer’s disease and brain tumor detection using deep learning with particle swarm optimization. AI. 2023;4(3):551–73. https://doi.org/10.3390/ai4030030.
Özkaraca O, Bağrıaçık Oİ, Gürüler H, Khan F, Hussain J, Khan J, Laila UE. Multiple brain tumor classification with dense CNN architecture using brain MRI images. Life. 2023;13(2):349. https://doi.org/10.3390/life13020349.
Noreen N, Palaniappan S, Qayyum A, Ahmad I, Imran M, Shoaib M. A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access. 2020;8:55135–44. https://doi.org/10.1109/ACCESS.2020.2978629.
Talukder MA, Islam MM, Uddin MA, Akhter A, Pramanik MAJ, Aryal S, Moni MA. An efficient deep learning model to categorize brain tumor using reconstruction and fine-tuning. Expert Syst Appl. 2023;230:120534. https://doi.org/10.1016/j.eswa.2023.120534.
Haq EU, Jianjun H, Li K, Haq HU, Zhang T. An MRI-based deep learning approach for efficient classification of brain tumors. J Ambient Intell Humaniz Comput. 2023. https://doi.org/10.1007/s12652-021-03535-9.
Malla PP, Sahu S, Alutaibi AI. Classification of tumor in brain MR images using deep convolutional neural network and global average pooling. Processes. 2023;11(3):679. https://doi.org/10.3390/pr11030679.
Rahman T, Islam MS. MRI brain tumor detection and classification using parallel deep convolutional neural networks. Meas Sens. 2023;26:100694. https://doi.org/10.1016/j.measen.2023.100694.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. ImageNet large scale visual recognition challenge. Int J Comput Vis. 115;3(December 2015):211–252. https://doi.org/10.1007/s11263-015-0816-y.
Shaw SS, Ahmed S, Malakar S, et al. Hybridization of ring theory-based evolutionary algorithm and particle swarm optimization to solve class imbalance problem. Complex Intell Syst. 2021;7:2069–91. https://doi.org/10.1007/s40747-021-00314-z.
Guha S, Das A, Singh PK, Ahmadian A, Senu N, Sarkar R. Hybrid feature selection method based on harmony search and naked mole-rat algorithms for spoken language identification from audio signals. IEEE Access. 2020;8:182868–87. https://doi.org/10.1109/ACCESS.2020.3028121.
Singh PK, Chatterjee I, Sarkar R. Page-level handwritten script identification using modified log-Gabor filter based features. In: 2015 IEEE 2nd international conference on recent trends in information systems (ReTIS), Kolkata, India, 2015. pp. 225–30. https://doi.org/10.1109/ReTIS.2015.7232882.
Reza A, Singh PK, Mahmud M, Brown DJ, Sarkar R. Multi-level stress detection using ensemble filter-based feature selection method. In: Mahmud M, Mendoza-Barrera C, Kaiser MS, Bandyopadhyay A, Ray K, Lugo E, editors. Proceedings of trends in electronics and health informatics. TEHI 2022. Lecture notes in networks and systems, vol. 675. Singapore: Springer. https://doi.org/10.1007/978-981-99-1916-1_13.
Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x.
Vergara J, Estevez P. A review of feature selection methods based on mutual information. Neural Comput Appl. 2014. https://doi.org/10.1007/s00521-013-1368-0.
Beraha M, Metelli AM, Papini M, Tirinzoni A, Restelli M. Feature selection via mutual information: new theoretical insights. In: 2019 international joint conference on neural networks (IJCNN), Budapest, Hungary, 2019. pp. 1–9. https://doi.org/10.1109/IJCNN.2019.8852410.
Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. https://doi.org/10.1007/BF00994018.
Boser B, Guyon I, Vapnik V. A training algorithm for optimal margin classifier. In: Proceedings of the fifth annual ACM workshop on computational learning theory, vol. 5. 1996. https://doi.org/10.1145/130385.130401.
Vapnik V. The nature of statistical learning theory. 2000. https://doi.org/10.1007/978-1-4757-3264-1_1.
Ghosh S, Dasgupta A, Swetapadma A. A study on support vector machine based linear and non-linear pattern classification. In: 2019 international conference on intelligent sustainable systems (ICISS), Palladam, India, 2019. pp. 24–8. https://doi.org/10.1109/ISS1.2019.8908018.
Bhuvaji S, Kadam A, Bhumkar P, Dedge S, Kanchan S. Brain tumor classification (MRI). Kaggle. 2020. https://doi.org/10.34740/KAGGLE/DSV/1183165.
Cheng J. Brain tumor dataset (Version 5). figshare. 2017. 10.6084/m9.figshare.1512427.v5.
Hamada A. Br35H:: brain tumor detection 2020 from Kaggle. 2020. https://www.kaggle.com/datasets/ahmedhamada0/brain-tumor-detection/data. Retrieved 20 Nov 2023.
Singh R, Prabha C. Enhancing accuracy in detection of glioma tumor using DenseNet CNN. In: 2023 international conference on research methodologies in knowledge management, artificial intelligence and telecommunication engineering (RMKMATE), Chennai, India, 2023. pp. 1–5. https://doi.org/10.1109/RMKMATE59243.2023.10369797.
Singh R, Prabha C, Kumari S, Murugan K, VMRM, Singh T. Accuracy enhancement in detecting pituitary tumors using deep learning. In: 2023 international conference on sustainable communication networks and application (ICSCNA), Theni, India, 2023. pp. 1067–72. https://doi.org/10.1109/ICSCNA58489.2023.10370139.
Waskita JM, Amda DSKS, Prasetio H. EfficientNetV2 based for MRI brain tumor image classification. In: 2023 international conference on computer, control, informatics and its applications (IC3INA), Bandung, Indonesia, 2023. pp. 171–6. https://doi.org/10.1109/IC3INA60834.2023.10285782.
Menze BH, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993–2024. https://doi.org/10.1109/TMI.2014.2377694.
Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4: 170117. https://doi.org/10.1038/sdata.2017.117.
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara R, Berger C, Ha S, Rozycki M, Prastawa M, Alberts E, Lipkova J, Freymann J, Kirby J, Bilello M, Fathallah-Shaykh H, Wiest R, Kirschke J, Chen, Zhaolin. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. 2019;38.
Schmainda KM, Prah M. Data from brain-tumor-progression. Cancer Imaging Arch. 2018. https://doi.org/10.7937/K9/TCIA.2018.15quzvnb.
Author information
Authors and Affiliations
Contributions
Conceptualization, P.K.S.; methodology, U.A.; software, P.K.S.; validation, S.K., U.A. and P.K.S.; formal analysis, U.A.; investigation, P.K.S.; resources, P.K.S.; data curation, S.K.; writing—original draft preparation, U.A.; writing—review and editing, P.K.S.; visualization, P.K.S., U.A.; supervision, P.K.S. and U.A.; project administration, P.K.S. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kar, S., Aich, U. & Singh, P.K. Efficient Brain Tumor Classification Using Filter-Based Deep Feature Selection Methodology. SN COMPUT. SCI. 5, 1033 (2024). https://doi.org/10.1007/s42979-024-03392-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-03392-1