Abstract
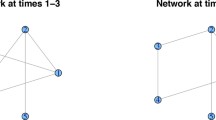
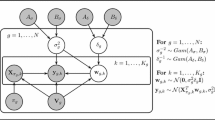
Recently, a Bayesian network model for inferring non-stationary regulatory processes from gene expression time series has been proposed. The Bayesian Gaussian Mixture (BGM) Bayesian network model divides the data into disjunct compartments (data subsets) by a free allocation model, and infers network structures, which are kept fixed for all compartments. Fixing the network structure allows for some information sharing among compartments, and each compartment is modelled separately and independently with the Gaussian BGe scoring metric for Bayesian networks. The BGM model can equally be applied to both static (steady-state) and dynamic (time series) gene expression data. However, it is this flexibility that renders its application to time series data suboptimal. To improve the performance of the BGM model on time series data we propose a revised approach in which the free allocation of data points is replaced by a changepoint process so as to take the temporal structure into account. The practical inference follows the Bayesian paradigm and approximately samples the network, the number of compartments and the changepoint locations from the posterior distribution with Markov chain Monte Carlo (MCMC). Our empirical results show that the proposed modification leads to a more efficient inference tool for analysing gene expression time series.
Similar content being viewed by others
References
Beal M, Falciani F, Ghahramani Z, Rangel C, Wild D (2005) A Bayesian approach to reconstructing genetic regulatory networks with hidden factors. Bioinformatics 21(3): 349–356
Darnell J, Kerr I, Stark G (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421
Friedman N, Koller D (2003) Being Bayesian about network structure. Mach Learn 50: 95–126
Geiger D, Heckerman D (1994) Learning Gaussian networks. Proceedings of the Tenth Conference on Uncertainty in Artificial Intelligence, pp 235–243
Gelman A, Rubin D (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7: 457–472
Green P (1995) Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82: 711–732
Grzegorczyk M, Husmeier D (2008) Improving the structure MCMC sampler for Bayesian networks by introducing a new edge reversal move. Mach Learn 71: 265–305
Grzegorczyk M, Husmeier D, Edwards K, Ghazal P, Millar A (2008) Modelling non-stationary gene regulatory processes with a non-homogeneous Bayesian network and the allocation sampler. Bioinformatics 24: 2071–2078
Heckerman D (1999) A tutorial on learning with Bayesian networks. In: Jordan MI (eds) Learning in graphical models, adaptive computation and machine learning. MIT Press, Cambridge, Massachusetts, pp 301–354
Honda K, Takaoka A, Taniguchi T. (2006) Type I interferon gene induction by the Interferon regulatory factor family of transcription factors. Immunity 25: 349–360
Husmeier D (2003) Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics 19: 2271–2282
Madigan D, York J (1995) Bayesian graphical models for discrete data. Int Stat Rev 63: 215–232
Raza S, Robertson K, Lacaze P, Page D, Enright A, Ghazal P, Freeman T (2008) A logic based diagram of signalling pathways central to macrophage activation. BMC Systems Biology 2:Article 36
Rogers S, Girolami M (2005) A Bayesian regression approach to the inference of regulatory networks from gene expression data. Bioinformatics 21(14): 3131–3137
Sachs K, Perez O, Pe‘er DA, Lauffenburger DA, Nolan GP (2005) Protein-signaling networks derived from multiparameter single-cell data. Science 308: 523–529
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grzegorczyk, M., Husmeier, D. & Rahnenführer, J. Modelling non-stationary dynamic gene regulatory processes with the BGM model. Comput Stat 26, 199–218 (2011). https://doi.org/10.1007/s00180-010-0201-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-010-0201-9