Abstract
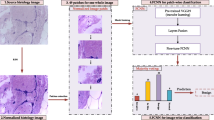
A large and balanced training data are the foremost requirement in proper convergence of a deep convolutional neural network (CNN). Medical data always suffer from the problem of unbalancing and inadequacy that makes it difficult to train CNN from scratch. It is known that the transfer learning approach provides great potential to deal with inadequate dataset besides the benefit of faster training. The efficient transfer of knowledge from natural images to histopathological images has yet to be achieved. In view of the foregoing, an attempt has been made toward the classification of BreakHis dataset using pre-trained ‘AlexNet’ model with a suitable fine-tuning approach. The effective depth of fine-tuning is also determined at different levels of magnification (40×, 100×, 200× and 400 ×). The experimental trials conform that the moderate level of fine-tuning is an optimum choice for the classification of magnification-dependent histology images in contrast to the shallow and deep tuning of the pre-trained network which in turn depends on the size and relative distribution of a dataset. Additionally, the layer-wise fine-tuning approach provides a neck-to-neck performance with the latest state-of-the-art developments.













Similar content being viewed by others
References
Tang, Z., Li, C., Sun, S.: Single-trial EEG classification of motor imagery using deep convolutional neural networks. Opt.-Int. J. Light Electron Opt. 130, 11–18 (2017)
Yang, Z., Mu, X.-D., Zhao, F.-A.: Scene classification of remote sensing image based on deep network and multi-scale features fusion. Optik 171, 287–293 (2018)
Zhang, J., Wang, C., Li, C., Qin, H.: Example-based rapid generation of vegetation on terrain via CNN-based distribution learning. Vis. Comput. 35(6–8), 1181–1191 (2019)
Sathish, D., Kamath, S., Prasad, K., Kadavigere, R.: Role of normalization of breast thermogram images and automatic classification of breast cancer. Visual Comput. 35(1), 57–70 (2019)
Soekhoe, D., van der Putten, P., Plaat, A.: On the impact of data set size in transfer learning using deep neural networks. In: International Symposium on Intelligent Data Analysis, pp. 50–60. Springer, Berlin (2016)
Wanderley, M.D.S., e Bueno, L.A., Zanchettin, C., Oliveira, A.L.: The impact of dataset complexity on transfer learning over convolutional neural networks. In: International Conference on Artificial Neural Networks 2017, pp. 582–589. Springer, Berlin
Yosinski, J., Clune, J., Bengio, Y., Lipson, H.: How transferable are features in deep neural networks? In: Advances in Neural Information Processing Systems 2014, pp. 3320–3328
Bar, Y., Diamant, I., Wolf, L., Greenspan, H.: Deep learning with non-medical training used for chest pathology identification. In: Medical Imaging 2015: Computer-Aided Diagnosis 2015, p. 94140V. International Society for Optics and Photonics
Tajbakhsh, N., Shin, J.Y., Gurudu, S.R., Hurst, R.T., Kendall, C.B., Gotway, M.B., Liang, J.: Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016)
Yang, H., Min, K.: Classification of basic artistic media based on a deep convolutional approach. In: The Visual Computer, pp. 1–20 (2019)
Tripathi, G., Singh, K., Vishwakarma, D.K.: Convolutional neural networks for crowd behaviour analysis: a survey. In: The Visual Computer, pp. 1–24 (2018)
Azizpour, H., Sharif Razavian, A., Sullivan, J., Maki, A., Carlsson, S.: From generic to specific deep representations for visual recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 36–45 (2015)
Margeta, J., Criminisi, A., Cabrera Lozoya, R., Lee, D.C., Ayache, N.: Fine-tuned convolutional neural nets for cardiac MRI acquisition plane recognition. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 5(5), 339–349 (2017)
Gao, M., Bagci, U., Lu, L., Wu, A., Buty, M., Shin, H.-C., Roth, H., Papadakis, G.Z., Depeursinge, A., Summers, R.M.: Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput. Methods Biomecha. Biomed. Eng. Imaging Vis. 6(1), 1–6 (2018)
Van Ginneken, B., Setio, A.A., Jacobs, C., Ciompi, F.: Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. In: IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 286–289. IEEE (2015)
Arevalo, J., González, F.A., Ramos-Pollán, R., Oliveira, J.L., Lopez, M.A.G.: Convolutional neural networks for mammography mass lesion classification. In: 37th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), pp. 797–800. IEEE (2015)
Carneiro, G., Nascimento, J., Bradley, A.P.: Unregistered multiview mammogram analysis with pre-trained deep learning models. In: International Conference on Medical Image Computing and Computer-Assisted Intervention 2015, pp. 652–660. Springer, Berlin
Chen, H., Ni, D., Qin, J., Li, S., Yang, X., Wang, T., Heng, P.A.: Standard plane localization in fetal ultrasound via domain transferred deep neural networks. IEEE J. Biomed. Health Inf. 19(5), 1627–1636 (2015)
Schlegl, T., Ofner, J., Langs, G.: Unsupervised pre-training across image domains improves lung tissue classification. In: International MICCAI Workshop on Medical Computer Vision, pp. 82–93. Springer, Berlin (2014)
Shin, H.-C., Lu, L., Kim, L., Seff, A., Yao, J., Summers, R.M.: Interleaved text/image deep mining on a very large-scale radiology database. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1090–1099 (2015)
Sharma, S., Mehra, R.: Automatic magnification independent classification of breast cancer tissue in histological images using deep convolutional neural network. In: International Conference on Advanced Informatics for Computing Research, pp. 772–781. Springer, Berlin (2018)
Alhindi, T.J., Kalra, S., Ng, K.H., Afrin, A., Tizhoosh, H.R.: Comparing LBP, HOG and deep features for classification of histopathology images. In: 2018 International Joint Conference on Neural Networks (IJCNN), pp. 1–7. IEEE (2018)
Wan, S., Huang, X., Lee, H.-C., Fujimoto, J.G., Zhou, C.: Spoke-LBP and ring-LBP: New texture features for tissue classification. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 195-199. IEEE (2015)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 63(7), 1455–1462 (2016)
Bengio, Y., Courville, A., Vincent, P.: Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1798–1828 (2013)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. nature 521(7553), 436 (2015)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: Breast cancer histopathological image classification using convolutional neural networks. In: 2016 International Joint Conference on Neural Networks (IJCNN), pp. 2560–2567. IEEE (2016)
Araújo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., Polónia, A., Campilho, A.: Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 12(6), e0177544 (2017)
Spanhol, F.A., Oliveira, L.S., Cavalin, P.R., Petitjean, C., Heutte, L.: Deep features for breast cancer histopathological image classification. In: 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), pp. 1868–1873. IEEE (2017)
Motlagh, N.H., Jannesary, M., Aboulkheyr, H., Khosravi, P., Elemento, O., Totonchi, M., Hajirasouliha, I.: Breast cancer histopathological image classification: A deep learning approach. bioRxiv, 242818 (2018)
Sharma, S., Mehra, R.: Breast cancer histology images classification: training from scratch or transfer learning? ICT Express 4(4), 247–254 (2018)
Han, Z., Wei, B., Zheng, Y., Yin, Y., Li, K., Li, S.: Breast cancer multi-classification from histopathological images with structured deep learning model. Sci Rep. 7(1), 4172 (2017)
Bardou, D., Zhang, K., Ahmad, S.M.: Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access 6, 24680–24693 (2018)
Chan, A., Tuszynski, J.A.: Automatic prediction of tumour malignancy in breast cancer with fractal dimension. R. Soc. Open Sci. 3(12), 160558 (2016)
Acknowledgements
The authors are immensely thankful to Dr. S.S Patnaik, Director, NITTTR, Chandigarh, for providing all the necessary facilities and support during the execution of the work. Also, the author Shallu Sharma would like to thank Dr. Sumit Kumar from IIT(ISM), Dhanbad, for every scientific discussion and constant support. The authors would like to thank Dr. Fabio Alexandre Spanhol for providing the BreakHis dataset.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, S., Mehra, R. Effect of layer-wise fine-tuning in magnification-dependent classification of breast cancer histopathological image. Vis Comput 36, 1755–1769 (2020). https://doi.org/10.1007/s00371-019-01768-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-019-01768-6