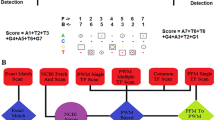
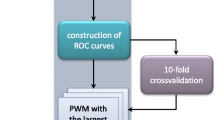
Abstract
Protein-DNA bindings are essential activities. Understanding them forms the basis for further deciphering of biological and genetic systems. In particular, the protein-DNA bindings between transcription factors (TFs) and transcription factor binding sites (TFBSs) play a central role in gene transcription. Comprehensive TF-TFBS binding sequence pairs have been found in a recent study. However, they are in one-to-one mappings which cannot fully reflect the many-to-many mappings within the bindings. An evolutionary algorithm is proposed to learn generalized representations (many-to-many mappings) from the TF-TFBS binding sequence pairs (one-to-one mappings). The generalized pairs are shown to be more meaningful than the original TF-TFBS binding sequence pairs. Some representative examples have been analyzed in this study. In particular, it shows that the TF-TFBS binding sequence pairs are not presumably in one-to-one mappings. They can also exhibit many-to-many mappings. The proposed method can help us extract such many-to-many information from the one-to-one TF-TFBS binding sequence pairs found in the previous study, providing further knowledge in understanding the bindings between TFs and TFBSs.









Similar content being viewed by others
Notes
As defined in Leung et al. (2010), a kmer is commonly found in a set of sequences if and only if it is a substring in more than or equal to half of the sequences.
A TFBS kmer–TF kmer pair is considered binding for a PDB chain if and only if an atom of the TFBS kmer and an atom of the TF kmer are close to each other. Two atoms are considered close if and only if their distance is smaller than 3.5 angstrom. Leung et al. (2010).
References
Aerts S, Van Loo P, Thijs G, Moreau Y, De Moor B (2003) Computational detection of cis-regulatory modules. Bioinformatics 19(Suppl 2):5–14
Agrawal R, Imielinski T, Swami A (1993) Mining association rules between sets of items in large databases. In: Proceedings of the 1993 ACM SIGMOD international conference on management of data, pp 207–216. doi:10.1145/170035.170072
Ahmad S, Gromiha MM, Sarai A (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics 20(4):477–486. doi:10.1093/bioinformatics/btg432
Ahmad S, Keskin O, Sarai A, Nussinov R (2008) Protein-DNA interactions: structural, thermodynamic and clustering patterns of conserved residues in DNA-binding proteins. Nucleic Acids Res 36:5922–5932
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Proceedings of the 2nd international conference on intelligent systems for molecular biology, pp 28–36
Bailey TL, Noble WS (2003) Searching for statistically significant regulatory modules. Bioinformatics 19(Suppl 2):16–25
Banzhaf W, Nordin P, Keller RE, Francone FD (1998) Genetic Programming—an introduction; on the automatic evolution of computer programs and its applications. Morgan Kaufmann, San Francisco
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, GrifRths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR (2004) The pfam protein families database. Nucleic Acids Res 32:D138–D141
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. doi:10.1093/nar/28.1.235
Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, Coulombe B, Robert F (2006) Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res 16:656–668
Brin S, Motwani R, Ullman JD, Tsur S (1997) Dynamic itemset counting and implication rules for market basket data. SIGMOD Rec 26(2):255–264. doi:10.1145/253262.253325
Coin L, Bateman A, Durbin R (2003) Enhanced protein domain discovery by using language modeling techniques from speech recognition. Proc Natl Acad Sci USA 100:4516–4520
Galas DJ, Schmitz A (1987) DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res 5(9):3157–3170
Garner MM, Revzin A (1981) A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the escherichia coli lactose operon regulatory system. Nucleic Acids Res 9(13):3047–3060
Givant S, Halmos P (2009) Introduction to boolean algebras. Springer, Berlin
Goldberg DE, Richardson J (1987) Genetic algorithms with sharing for multimodal function optimization. In: Proceedings of the 2nd international conference on genetic algorithms and their application. L. Erlbaum Associates Inc., Hillsdale, pp 41–49
Grundy WN, Bailey TL, Elkan CP, Baker ME (1997)Meta-MEME: motif-based hidden Markov models of protein families. Comput Appl Biosci 13:397–406
Holland JH (1975) Adaptation in natural and artificial systems. University of Michigan Press, Ann Arbor
Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJA (2008) The 20 years of prosite. Nucl Acids Res 36(Suppl 1):D245–D249
Jensen ST, Liu XS, Zhou Q, Liu JS (2004) Computational discovery of gene regulatory binding motifs: a bayesian perspective. Stat Sci 19(1):188–204
Jong KAD (1975) An analysis of the behavior of a class of genetic adaptive systems. PhD thesis, University of Michigan, Ann Arbor
Jong KAD (2006) Evolutionary Computation. A Unified Approach. MIT Press, Cambridge, MA
Karnaugh M (1953) A map method for synthesis of combinational logic circuits. Trans AIEE Commun Electron 72 (I):593–599
Kato M, Hata N, Banerjee N, Futcher B, Zhang MQ (2004) Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol 5:R56
Kel-Margoulis OV, Kel AE, Reuter I, Deineko IV, Wingender E (2002) TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res 30:332–334
Kraft D, Petry F, Buckles B, Sadasivan T (1994) The use of genetic programming to build queries for information retrieval. In: Evolutionary Computation, 1994. IEEE World Congress on Computational Intelligence. Proceedings of the 1st IEEE conference, vol 1, pp 468–473. doi:10.1109/ICEC.1994.349905
Krivan W, Wasserman WW (2001) A predictive model for regulatory sequences directing liver-specific transcription. Genome Res 11:1559–1566
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Leung KS, Wong KC, Chan TM, Wong MH, Lee KH, Lau CK, Tsui SKW (2010) Discovering protein-DNA binding sequence patterns using association rule mining. Nucleic Acids Research (accepted)
Li JP, Balazs ME, Parks GT, Clarkson PJ (2002) A species conserving genetic algorithm for multimodal function optimization. Evol Comput 10(3):207–234. doi:10.1162/106365602760234081
Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein-DNA binding sites with applications to chromatinimmunoprecipitation microarray experiments. Nat Biotechnol 20:835–839
Luscombe NM, Thornton JM (2002) Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J Mol Biol 320(5):991–1009
Luscombe NM, Austin SE, Berman HM, Thornton JM (2000) An overview of the structures of protein-DNA complexes. Genome Biol 1(1):1–37
MacIsaac KD, Fraenkel E (2006) Practical strategies for discovering regulatory DNA sequence motifs. PLoS Comput Biol 2(4):e36
Matys V, Kel-Margoulis O, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel A, Wingender E (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34:D108–D110
McGuire AM, De Wulf P, Church GM, Lin EC (1999) A weight matrix for binding recognition by the redox-response regulator ArcA-P of Escherichia coli. Mol Microbiol 32:219–221
Mohan PM, Hosur RV (2009) Structure-function-folding relationships and native energy landscape of dynein light chain protein: nuclear magnetic resonance insights. J Biosci 34:465–479
Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE (2005) The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics 6:21
Nelson RJ (1953) A way to simplify truth functions. J Symb Logic 18(3):280–282
Nelson VP, Nagle HT, Carroll BD, Irwin JD (1995) Digital logic circuit analysis and design. Prentice-Hall, Inc., Upper Saddle River
Ofran Y, Mysore V, Rost B (2007) Prediction of DNA-binding residues from sequence. Bioinformatics 23(13):i347–i353. doi:10.1093/bioinformatics/btm174
Pavlidis P, Furey TS, Liberto M, Haussler D, Grundy WN (2001) Promoter region-based classification of genes. In: Pacific symposium on biocomputing, pp 151–163
Remenyi A, Scholer HR, Wilmanns M (2004) Combinatorial control of gene expression. Nat Struct Mol Biol 11:812–815
Rudell RL (1986) Multiple-valued logic minimization for pla synthesis. Tech. Rep. UCB/ERL M86/65, EECS Department, University of California, Berkeley. http://www.eecs.berkeley.edu/Pubs/TechRpts/1986/734.html
Smith AD, Sumazin P, Das D, Zhang MQ (2005) Mining ChIP-chip data for transcription factor and cofactor binding sites. Bioinformatics Suppl 1(20):i403–i412
Smyth MS, Martin JH (2000) X-ray crystallography. Mol Pathol 53(1):8–14
Stormo GD (1988) Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev BioChem 17:241–263
Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD (2008) The evolution of combinatorial gene regulation in fungi. PLoS Biol 6:e38
Veitch EW (1952) A chart method for simplifying truth functions. In: Proceedings of the 1952 ACM national meeting, Pittsburgh. ACM, New York, pp 127–133. doi:10.1145/609784.609801
Wegner M (1999) From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res 27:1409–1420
White RJ (2001) Gene transcription: mechanisms and control. Blackwell, Oxford
Wolberger C (1998) Combinatorial transcription factors. Curr Opin Genet Dev 8:552–559
Wong KC, Leung KS, Wong MH (2009) An evolutionary algorithm with species-specific explosion for multimodal optimization. In: Proceedings of the 11th Annual conference on genetic and evolutionary computation. ACM, New York, pp 923–930. doi:10.1145/1569901.1570027
Wong KC, Leung KS, Wong MH (2010a) Effect of spatial locality on an evolutionary algorithm for multimodal optimization. In: Applications of Evolutionary Computation, EvoApplications 2010 Part I. Lecture notes in computer science, vol 6024. Springer, Berlin, pp 481–490. doi:10.1007/978-3-642-12239-2_50
Wong KC, Leung KS, Wong MH (2010b) Protein structure prediction on a lattice model via multimodal optimization techniques. In: Proceedings of the 12th annual conference on genetic and evolutionary computation. ACM, New York, pp 155–162. doi:10.1145/1830483.1830513
Zhou Q, Liu JS (2008) Extracting sequence features to predict protein-DNA interactions: a comparative study. Nucleic Acids Res 36(12):4137–4148. doi:10.1093/nar/gkn361
Acknowledgments
The authors are grateful to the anonymous reviewers for their valuable comments. They would like to thank Tak-Ming Chan for his help on surveying the related works. This research is partially supported by the grants from the Research Grants Council of the Hong Kong Special Administrative Region, China (Project Nos. 414107 and 414708).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wong, KC., Peng, C., Wong, MH. et al. Generalizing and learning protein-DNA binding sequence representations by an evolutionary algorithm. Soft Comput 15, 1631–1642 (2011). https://doi.org/10.1007/s00500-011-0692-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-011-0692-5