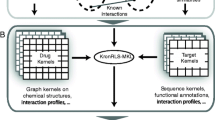
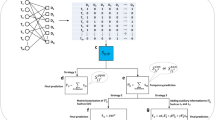
Abstract
With the emergence of large-scale experimental data on genes and proteins, drug discovery and repositioning will be more difficult in the field of biomedical research. More and more resources are needed for detecting drug–target interactions (DTIs) in the experimental works. The interactions between drugs and targets could been seen as a bipartite network. Many computational methods have been developed to identify DTIs. However, most of them did not integrate multiple information and filter noise or outlier points. In this paper, we develop a fuzzy bipartite local model (FBLM) based on fuzzy least squares support vector machine and multiple kernel learning (MKL) for predicting DTIs. First, multiple kernels are constructed in drug and target spaces, respectively. Then, all corresponding kernels are combined by MKL algorithm in two spaces. Finally, FBLM is employed to identify DTIs. Our proposed approach is tested on four benchmark datasets under three types of cross validation. Comparing with existing outstanding methods, our method is a useful tool for the DTIs prediction.








Similar content being viewed by others
References
Hecker N, Ahmed J, Von Eichborn J et al (2012) SuperTarget goes quantitative: update on drug–target interactions. Nucleic Acids Res 40(D1):1113–1117
Schomburg I, Chang A, Placzek S et al (2013) Brenda in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in brenda. Nucleic Acids Res 41(D1):764–772
Kanehisa M, Goto S, Hattori M et al (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34(Suppl 1):354–357
Law V, Knox C, Djoumbou Y et al (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42(D1):1091–1097
Chen X, Liu MX, Yan GY (2012) Drug–target interaction prediction by random walk on the heterogeneous network. Mol BioSyst 8(7):1970–1978
Feixiong C, Chuang L, Jing J et al (2012) Prediction of drug–target interactions and drug repositioning via network-based inference. PLoS Comput Biol 8(5):e1002503
Lin C, Chen W, Qiu C et al (2014) LibD3C: ensemble classifiers with a clustering and dynamic selection strategy. Neurocomputing 123:424–435
Ding YJ, Tang JJ, Guo F (2017) Identification of drug–target interactions via multiple information integration. Inf Sci 418:546–560
Ding YJ, Tang JJ, Guo F (2019) Identification of drug-side effect association via multiple information integration with centered kernel alignment. Neurocomputing 325:211–224
Qu W, Yang B, Jiang W et al (2012) HYBP-PSSP: a hybrid back propagation method for predicting protein secondary structure. Neural Comput Appl 21(2):337–349
Dongardive J, Abraham S (2017) Reaching optimized parameter set: protein secondary structure prediction using neural network. Neural Comput Appl 28(8):1947–1974
Wang YC, Zhang CH, Deng NY et al (2011) Research article: kernel-based data fusion improves the drug–protein interaction prediction. Comput Biol Chem 35(6):353–362
Peng LH, Liao B, Zhu W, Li ZJ, Li KQ (2015) Predicting drug–target interactions with multi-information fusion. IEEE J Biomed Health Inf 21(2):561–572
Bleakley K, Yamanishi Y (2009) Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics 25(18):2397–2403
Mei JP, Kwoh CK, Yang P et al (2013) Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics 29(2):238–245
Xia Z, Wu LY, Zhou X et al (2010) Semi-supervised drug–protein interaction prediction from heterogeneous biological spaces. BMC Syst Biol 4(Suppl 2):6–17
Van LT, Marchiori E (2013) Predicting drug–target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS ONE 8(6):e66952(1)–e66952(6)
Van LT, Nabuurs SB, Marchiori E (2011) Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 27(21):3036–3043
Cichonska A, Ravikumar B, Parri E et al (2017) Computational–experimental approach to drug–target interaction mapping: a case study on kinase inhibitors. Plos Comput Biol 13(8):e1005678
Hao M, Wang Y, Bryant SH (2016) Improved prediction of drug–target interactions using regularized least squares integrating with kernel fusion technique. Anal Chim Acta 909:41–50
Nascimento ACA, Prudêncio RBC, Costa IG (2016) A multiple kernel learning algorithm for drug–target interaction prediction. BMC Bioinform 17(1):46
Zhang W, Chen Y, Li D (2017) Drug–target interaction prediction through label propagation with linear neighborhood information. Molecules 22(12):2056–2069
Zheng X, Ding H, Mamitsuka H et al (2013) Collaborative matrix factorization with multiple similarities for predicting drug-target interactions. In: ACM SIGKDD international conference on knowledge discovery and data mining, pp 1025–1033
Gönen M (2012) Predicting drug–target interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics 28(18):2304–2310
Liu Y, Wu M, Miao C et al (2016) Neighborhood regularized logistic matrix factorization for drug–target interaction prediction. Plos Comput Biol 12(2):e1004760
Ezzat A, Zhao P, Wu M et al (2016) Drug–target interaction prediction with graph regularized matrix factorization. IEEE-ACM Trans Comput Biol Bioinform 14(3):646–656
Bolgar B, Antal P (2017) VB-MK-LMF: fusion of drugs, targets and interactions using variational Bayesian multiple kernel logistic matrix factorization. BMC Bioinform 18(1):440–457
Luo Y, Zhao X, Zhou J et al (2017) A network integration approach for drug–target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun. https://doi.org/10.1038/s41467-017-00680-8
Mousavian Z, Khakabimamaghani S, Kavousi K et al (2015) Drug–target interaction prediction from PSSM based evolutionary information. J Pharmacol Toxicol Methods 78:42–51
Cao DS, Liu S, Xu QS et al (2012) Large-scale prediction of drug–target interactions using protein sequences and drug topological structures. Anal Chim Acta 752(21):1–10
Cao DS, Zhang LX, Tan GS et al (2014) Computational prediction of drug target interactions using chemical, biological, and network features. Mol Inform 33(10):669–681
Li Z, Han P, You Z et al (2017) In silico prediction of drug–target interaction networks based on drug chemical structure and protein sequences. Sci Rep 7(1):11174
Gui J, Liu T, Tao D et al (2017) Representative vector machines: a unified framework for classical classifiers. IEEE Trans Cybern 46(8):1877–1888
Kurgan L, Wang C (2018) Survey of similarity-based prediction of drug–protein interactions. Curr Med Chem. https://doi.org/10.2174/0929867325666181101115314
Zhou L, Li Z, Yang J et al (2019) Revealing drug–target interactions with computational models and algorithms. Molecules 24(9):1714
Ezzat A, Wu M, Li X et al (2019) Computational prediction of drug–target interactions via ensemble learning. Methods Mol Biol. https://doi.org/10.1007/978-1-4939-8955-3-14
Lee I, Keum J, Nam H (2019) DeepConv-DTI: prediction of drug–target interactions via deep learning with convolution on protein sequences. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1007129
Hattori M, Okuno Y, Goto S et al (2003) Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J Am Chem Soc 125(39):11853–11865
Takarabe M, Kotera M, Nishimura Y et al (2012) Drug target prediction using adverse event report systems. Bioinformatics 28(18):i611–i618
Yamanishi Y, Araki M, Gutteridge A et al (2008) Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 24(13):i232–i240
Smedley D, Haider S, Durinck S et al (2015) The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 43(1):589–598
Ovaska K, Laakso M, Hautaniemi S (2008) Fast gene ontology based clustering for microarray experiments. Biodata Min 1(1):11–11
Perlman L, Gottlieb A, Atias N et al (2011) Combining drug and gene similarity measures for drug–target elucidation. J Comput Biol A J Comput Mol Cell Biol 18(2):133
He J, Chang SF, Xie L (2008) Fast kernel learning for spatial pyramid matching. In: IEEE conference on computer vision and pattern recognition
Tsujinishi D, Abe S (2003) Fuzzy least squares support vector machines for multiclass problems. Neural Netw 16(5–6):785–792
Tsujinishi D, Abe S (2003) Fuzzy least squares support vector machines. Proc 2003 Int Jt Conf Neural Netw 2:1599–1604
Suykens JAK, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Process Lett 9(3):293–300
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297
Lin CF, Wang SD (2002) Fuzzy support vector machines. IEEE Trans Neural Netw 13(2):464–471
Gaulton A, Bellis LJ, Bento AP et al (2012) Chembl: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40(DI):D1100–D1107
Günther S, Kuhn M, Dunkel M et al (2008) SuperTarget and matador: resources for exploring drug–target relationships. Nucleic Acids Res 36:919–922
Acknowledgements
This work is supported by a grant from the National Science Foundation of China (NSFC 61772362 and 61902271), Natural Science Research Project of Jiangsu Higher Eduction Institutions of China (19KJB520014) and the Tianjin Research Program of Application Foundation and Advanced Technology (16JCQNJC00200). The authors also thank Dr. Yamanishi Y., Liu Y. and Nascimento A.C.A. for kindly providing datasets on their websites.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Availability of data and material
The source code, datasets and corresponding results are available at https://figshare.com/s/837b5e6631ed95dfc94e.
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ding, Y., Tang, J. & Guo, F. Identification of drug–target interactions via fuzzy bipartite local model. Neural Comput & Applic 32, 10303–10319 (2020). https://doi.org/10.1007/s00521-019-04569-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-019-04569-z