Abstract
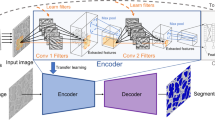
While the science and technology of nanostructure have rapidly been developed in many fields, it is still hard to obtain sufficient samples of nano objects due to high cost, thus, impeding the development of deep learning approaches to the material fields. Here, we develop a novel approach to recognize nano objects in Atomic Force Microscope (AFM) images. First, a noise reduction method based on the Laplacian of the Gaussian(LoG) is represented to denoise the AFM images. Then, two improved methods based on the watershed algorithm are proposed to segment the overlapping objects. Finally, a CNN recognition model based on transfer learning which is pre-trained on a large scale of shapes of handwritten numbers and letters is built to recognize the nano objects in AFM images. These methods can resolve effectively the small sample problem of AFM image processing.














Similar content being viewed by others
References
François P, De FAF, Menachem E (2015) Environmental applications of graphene-based nanomaterials. Chem Soc Rev 44(16):5861–5896
Nan G, Xiaosheng F (2015) Synthesis and development of grapheneinorganic semiconductor nanocomposites. Chem Rev 115(16):8294–8343
Yanmei S, Bin Z (2016) Recent advances in transition metal phosphide nanomaterials: synthesis and applications in hydrogen evolution reaction. Chem Soc Rev 45(6):1529–1541
Qilin L, Shaily M, Lyon Delina Y, Lena B, Liga Michael V, Dong L, Alvarez Pedro JJ (2008) Antimicrobial nanomaterials for water disinfection and microbial control: potential applications and implications. Water Res 42(18):4591–4602
Shuilin W, Zhengyang W, Xiangmei Liu KWK, Chu YP et al (2014) Functionalized tio2 based nanomaterials for biomedical applications. Adv Funct Mater 24(35):5464–5481
Yin Zhang TR, Hong NH, Weibo C (2013) Biomedical applications of zinc oxide nanomaterials. Curr Mol Med 13(10):1633–1645
Guohai Y, Chengzhou Z, Dan D, Junjie Z, Yuehe L (2015) Graphene-like two-dimensional layered nanomaterials: applications in biosensors and nanomedicine. Nanoscale 7(34):14217–14231
Tour James M (2013) Top-down versus bottom-up fabrication of graphene-based electronics. Chem Mater 26(1):163–171
Satoshi M, Akihiko K, Satoshi K, Hirohide S, Masami H (2013) Molecular robotics: a new paradigm for artifacts. New Gener Comput 31(1):27–45
Yuexing H, Akito H, Akinori K, Ryosuke W, Yuichi O, Akihiko K (2015) Automatic recognition of dna pliers in atomic force microscopy images. New Gener Comput 33(3):253–270
Haojun W, Chongxun Z, Dong M (2001) Applying watershed algorithm to segmentation of color images from blood and bone marrow smears. J Xian Jiaotong Univ 35(12):1296–1299
Boray Tek F, Dempster Andrew G, Izzet Kale (2005) Blood cell segmentation using minimum area watershed and circle radon transformations. Springer, Netherlands
Jierong C, Rajapakse Jagath C et al (2009) Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng 56(3):741–748
Qian W, Yuexing H, Qing L, Bing W, Akihiko K (2018) Segmenting overlapping nano-objects in atomic force microscopy image. J Nanophoton 12(1):016003
Huaibo S, Chuandong Z, Jingpeng P, Yibin Z, Xu Y (2013) Segmentation and reconstruction of overlapped apple images based on convex hull. Trans Chin Soc Agric Eng 29(3):163–168
Chanho J, Changick K, Wan CS, Sukjoong O (2010) Unsupervised segmentation of overlapped nuclei using bayesian classification. IEEE Trans Biomed Eng 57(12):2825–2832
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22(8):888–905
Felzenszwalb Pedro F, Huttenlocher Daniel P (2004) Efficient graph-based image segmentation. Int J Comput Vis 59(2):167–181
Ondřej Daněk, Pavel Matula, Carlos Ortiz-de Solórzano, Arrate Muñoz-Barrutia, Martin Maška, Michal Kozubek (2009) Segmentation of touching cell nuclei using a two-stage graphcut model. In: Scandinavian conference on image analysis pp. 410–419
Chiwoo P, Huang JZ, Ji JX, Ding Y (2012) Segmentation, inference and classification of partially overlapping nanoparticles. IEEE Trans Pattern Anal Mach Intell 35(3):1–1
Christoph S, Gerlich DW (2013) Machine learning in cell biology-teaching computers to recognize phenotypes. J Cell Sci 126(24):5529–5539
François C (2017) Xception: deep learning with depthwise separable convolutions. In: proceedings of the IEEE conference on computer vision and pattern recognition pp. 1251–1258
Tsung-Han C, Kui J, Shenghua G, Jiwen L, Zinan Z, Yi M (2015) Pcanet: a simple deep learning baseline for image classification. IEEE Trans Image Process 24(12):5017–5032
Jialin PS, Qiang Y (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Dan CC, U Meier, J Schmidhuber (2012) Transfer learning for latin and chinese characters with deep neural networks. In: the 2012 international joint conference on neural networks (IJCNN), pp. 1–6
Akinori K, Yusuke S, Takahiro YX, Komiyama YM (2011) Nanomechanical dna origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat Commun 2(1):449
Neves João C, Helena C, Ana T, Miguel C, Hugo P (2014) Detection and separation of overlapping cells based on contour concavity for leishmania images. Cytom Part A 85(6):491–500
Tony Lindeberg (1994) Scale-space theory in computer vision. Kluwer Academic, Netherlands
Nobuyuki O (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cyber 9(1):62–66
Malpica N, de Solórzano C, Vaquero JJ, Santos A, Vallcorba I, García-Sagredo JM, Del Pozo F (2015) Applying watershed algorithms to the segmentation of clustered nuclei. Cytom Part B Clin Cytom 28(4):289–297
Al Hamad HA, Zitar RA (2010) Development of an efficient neural-based segmentation technique for arabic handwriting recognition. Pattern Recog 43(8):2773–2798
Lindeman MA (2000) Influences of land use on the biogeochemistry of streams. Fundamenta Informaticae 41(1–2):187–228
Ron K, Nahum K, Bruckstein AM (1996) Sub-pixel distance maps and weighted distance transforms. J Math Imaging Vis 6(2–3):223–233
Granlund Gösta H (1972) Fourier preprocessing for hand print character recognition. IEEE Trans Comput 100(2):195–201
Jacob F, Manish S (2005) Information along contours and object boundaries. Psychol Rev 112(1):243
Corinna C, Vladimir V (1995) Support-vector networks. Mach Learn 20(3):273–297
Irina Rish et al (2001) An empirical study of the naive bayes classifier. In: IJCAI 2001 workshop on empirical methods in artificial intelligence vol 3, pp. 41–46
Kai HL, Peter S (1990) Neural network ensembles. IEEE Trans Pattern Anal Mach Intell 12(10):993–1001
Xuhong L, Yves G, Franck D (2020) A baseline regularization scheme for transfer learning with convolutional neural networks. Pattern Recog 98:107049
Zhang X, Zou J, He K, Sun J (2015) Accelerating very deep convolutional networks for classification and detection. IEEE Trans Pattern Anal Mach Intell 38(10):1943–1955
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: proceedings of the IEEE conference on computer vision and pattern recognition pp. 770–778
Iandola F, Moskewicz M, Karayev S, Girshick R, Darrell T, Keutzer K (2014) Densenet: implementing efficient convnet descriptor pyramids. arXiv preprint arXiv:1404.1869
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition 2818–2826
Acknowledgements
This research is sponsored by the National Key Research and Development Program of China (Grant Nos. 2018YFB0704400, 2018YFB0704402, 2020YFB0704503), Natural Science Foundation of Shanghai (Grant No. 20ZR1419000).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no financial and personal conflict of Interest statement with any person or organizations that could inappropriately influence our work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Han, Y., Liu, Y., Wang, B. et al. A novel transfer learning for recognition of overlapping nano object. Neural Comput & Applic 34, 5729–5741 (2022). https://doi.org/10.1007/s00521-021-06731-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-06731-y