Abstract
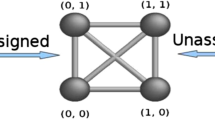
Considering geometry based on the concept of distance, the results found by Menger and Blumenthal originated a body of knowledge called distance geometry. This survey covers some recent developments for assigned and unassigned distance geometry and focuses on two main applications: determination of three-dimensional conformations of biological molecules and nanostructures.















Similar content being viewed by others
References
Almeida FCL, Moraes AH, Gomes-Neto F (2013) An overview on protein structure determination by NMR. In: Mucherino A et al (eds) Historical and future perspectives of the use of distance geometry methods, pp 377–412
Berger B, Kleinberg J, Leighton T (2011) Reconstructing a three-dimensional model with arbitrary errors. J Assoc Comput Mach 50:212–235
Billinge SJL (2010) Viewpoint: the nanostructure problem. Physics 3:25
Billinge SJL, Kanatzidis MG (2004) Beyond crystallography: the study of disorder, nanocrystallinity and crystallographically challenged materials with pair distribution functions. Chem Commun 7:749–760
Billinge SJL, Levin I (2007) The problem with determining atomic structure at the nanoscale. Science 316(5824):561–565
Blumenthal LM (1953) Theory and applications of distance geometry. Oxford University Press, Oxford
Bouchevreau B, Martineau C, Mellot-Draznieks C, Tuel A, Suchomel MR, Trebosc J, Lafon O, Amoureux JP, Taulelle F (2013) An NMR-driven crystallography strategy to overcome the computability limit of powder structure determination: A layered aluminophosphate case. Int J Comput Geom Appl 19:5009–5013
Boutin M, Kemper G (2007) Which point configurations are determined by the distribution of their pairwise distances. Int J Comput Geom Appl 17(1):31–43
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr 54(595):905–921
Carvalho RS, Lavor C, Protti F (2008) Extending the geometric build-up algorithm for the molecular distance geometry problem. Inf Process Lett 108:234–237
Cassioli A, Bardiaux B, Bouvier G, Mucherino A, Alves R, Liberti L, Nilges M, Lavor C, Malliavin TE (2015a) An algorithm to enumerate all possible protein conformations verifying a set of distance restraints. BMC Bioinform 16:23
Cassioli A, Gunluk O, Lavor C, Liberti L (2015b) Discretization vertex orders in distance geometry. Discrete Appl Math 197:27–41
Clore GM, Gronenborn AM (1997) New methods of structure refinement for macromolecular structure determination by NMR. Proc Natl Acad Sci 95:5891–5898
Connelly R (1991) On generic global rigidity. DIMACS Ser Discrete Math Theor Comput Sci 4:147–155
Connelly R (2005) Generic global rigidity. Discrete Comput Geom 33:549–563
Connelly R (2013) Generic global rigidity of body-bar frameworks. J Comb Theory Ser B 103:689–705
Costa V, Mucherino A, Lavor C, Cassioli A, Carvalho L, Maculan N (2014) Discretization orders for protein side chains. J Glob Optim 60:333–349
Crippen GM, Havel TF (1988) Distance geometry and molecular conformation. Research Studies Press, Baldock
Dokmanic I, Parhizkar R, Ranieri J, Vetterli M (2015) Euclidean distance matrices: essential theory, algorithms, and applications. IEEE Signal Process Mag 32(6):12–30
Dong Q, Wu Z (2002) A linear-time algorithm for solving the molecular distance geometry problem with exact interatomic distances. J Glob Optim 22:365–375
Duxbury PM, Granlund L, Gujarathi SR, Juhas P, Billinge SJL (2016) The unassigned distance geometry problem. Discrete Appl Math 204:117–132
Egami T, Billinge SJL (2012) Underneath the Bragg peaks: structural analysis of complex materials, 2nd edn. Pergamon Press, Oxford
Eren T, Goldenberg DK, Whiteley W, Yang YR, Morse AS, Anderson BDO, Belhumeur PN (2004) Rigidity, computation and randomization in network localization. In: 23rd annual joint conference of the IEEE computer and communications societies, vol 4, pp 2673–2684
Evrard G, Pusztai L (2005) Reverse Monte Carlo modelling of the structure of disordered materials with RMC++: a new implementation of the algorithm in C++. J Phys Condens Matter 17:S1–S13
Farrow CL, Juhas P, Liu JW, Bryndin D, Boz̈in ES, Bloch J, Proffen T, Billinge SJL (2007) PDFfit2 and PDFgui: computer programs for studying nanostructure in crystals. J Phys Condens Matter 19(33):335219
Gaffney KJ, Chapman HN (2007) Imaging atomic structure and dynamics with ultrafast X-ray scattering. Science 36(5830):1444–1448
Gommes CJ, Jiao Y, Torquato S (2012) Microstructural degeneracy associated with a two-point correlation function and its information contents. Phys Rev E 85:051140
Gonçalves D, Mucherino A (2014) Discretization orders and efficient computation of Cartesian coordinates for distance geometry. Optim Lett 8:2111–2125
Gonçalves DS, Mucherino A, Lavor C (2014) An adaptive branching scheme for the branch & prune algorithm applied to distance geometry. In: IEEE conference proceedings, federated conference on computer science and information systems (FedCSIS 14), workshop on computational optimization (WCO14), Warsaw, Poland, pp 463–469
Gortler S, Healy A, Thurston D (2010) Characterizing generic global rigidity. Am J Math 132(4):897–939
Graver J, Servatius B, Servatius H (1993) Combinatorial rigidity. American Mathematical Society, issue 2 of graduate studies in mathematics
Guerry P, Herrmann T (2011) Advances in automated NMR protein structure determination. Q Rev Biophys 44(3):257–309
Gujarathi SR, Farrow CL, Glosser C, Granlund L, Duxbury PM (2014) Ab-initio reconstruction of complex Euclidean networks in two dimensions. Phys Rev 89:053311
Hendrickson B (1992) Conditions for unique graph realizations. SIAM J Comput 21:65–84
Hendrickson B (1995) The molecule problem: exploiting structure in global optimization. SIAM J Optim 5(4):835–857
Jackson B, Jordan T (2005) Connected rigidity matroids and unique realization graphs. J Comb Theory Ser B 94:1–29
Jacobs DJ, Hendrickson B (1997) An algorithm for two-dimensional rigidity percolation: the pebble game. J Comput Phys 137:346–365
Jacobs DJ, Thorpe MF (1995) Generic rigidity percolation: the pebble game. Phys Rev Lett 75(22):4051–4054
Jaganathan K, Hassibi B (2013) Reconstruction of integers from pairwise distances. In: ICASSP, pp 5974–5978
Jain PC, Trigunayat GC (1977) Resolution of ambiguities in Zhdanov notation: actual examples of homometric structures. Acta Crystallogr A33:257–260
Juhás P, Cherba DM, Duxbury PM, Punch WF, Billinge SJL (2006) Ab initio determination of solid-state nanostructure. Nature 440(7084):655–658
Juhás P, Granlund L, Duxbury PM, Punch WF, Billinge SJL (2008) The LIGA algorithm for ab initio determination of nanostructure. Acta Crystallogr Sect A Found Crystallogr 64(Pt 6):631–640
Juhas P, Granlund L, Gujarathi SR, Duxbury PM, Billinge SJL (2010) Crystal structure solution from experimentally determined atomic pair distribution functions. J Appl Crystallogr 43:623–629
Laman G (1970) On graphs and rigidity of plane skeletal structures. J Eng Math 4:331–340
Lavor C, Mucherino A, Liberti L, Maculan N (2011) On the computation of protein backbones by using artificial backbones of hydrogens. J Glob Optim 50:329–344
Lavor C, Lee J, Lee-St.John A, Liberti L, Mucherino A, Sviridenko M (2012a) Discretization orders for distance geometry problems. Optim Lett 6(4):783–796
Lavor C, Liberti L, Maculan N, Mucherino A (2012b) The discretizable molecular distance geometry problem. Comput Optim Appl 52:115–146
Lavor C, Liberti L, Mucherino A (2013) The interval BP algorithm for the discretizable molecular distance geometry problem with interval data. J Glob Optim 56:855–871
Lavor C, Alves R, Figueiredo W, Petraglia A, Maculan N (2015) Clifford algebra and the discretizable molecular distance geometry problem. Adv Appl Clifford Algebras 25:925–942
Liberti L, Lavor C (2015) Six mathematical gems from the history of distance geometry. Int Trans Oper Res. doi:10.1111/itor.12170
Liberti L, Lavor C, Mucherino A (2013) In Mucherino A et al (eds) The discretizable molecular distance geometry problem seems easier on proteins, pp 47–60
Liberti L, Lavor C, Maculan N, Mucherino A (2014a) Euclidean distance geometry and applications. SIAM Rev 56(1):3–69
Liberti L, Masson B, Lee J, Lavor C, Mucherino A (2014b) On the number of realizations of certain Henneberg graphs arising in protein conformation. Discrete Appl Math 165:213–232
Lovász L, Yemini Y (1982) On generic rigidity in the plane. SIAM J Algorithms Discrete Math 3:91–98
McGreevy RL, Pusztai L (1988) Reverse Monte Carlo simulation: a new technique for the determination of disordered structures. Mol Simul 1:359–367
Menger K (1928) Dimension theorie. Teubner, Berlin
Moukarzel C (1996) An efficient algorithm for testing the generic rigidity of graphs in the plane. J Phys A Math Gen 29:8079–8098
Moukarzel C, Duxbury PM (1995) Stressed backbone and elasticity of random central-force system. Phys Rev Lett 75(22):4055–4058
Mucherino A (2013) On the identification of discretization orders for distance geometry with intervals. In: Nielsen F, Barbaresco F (eds) Proceedings of geometric science of information (GSI 13). Lecture Notes in Computer Science, vol 8085, Paris, France, pp 231–238
Mucherino A (2015) A pseudo de bruijn graph representation for discretization orders for distance geometry. In: Ortuño F, Rojas I (eds) Lecture Notes in Computer Science, vol 9043, Lecture Notes in Bioinformatics series, Proceedings of the 3rd international work-conference on bioinformatics and biomedical engineering (IWBBIO15), Granada, Spain, pp 514–523
Mucherino A, Lavor C, Malliavin T, Liberti L, Nilges M, Maculan N (2011) Influence of pruning devices on the solution of molecular distance geometry problems. In: Pardalos PM, Rebennack S (eds) Lecture Notes in Computer Science, vol 6630, Proceedings of the 10th international symposium on experimental algorithms (SEA11), Crete, Greece, pp 206–217
Mucherino A, Lavor C, Liberti L (2012) The discretizable distance geometry problem. Optim Lett 6:1671–1686
Mucherino A, Lavor C, Liberti L, Maculan N (eds) (2013) Distance geometry: theory, methods, and applications. Springer, Berlin
Mucherino A, de Freitas R, Lavor C (2015) Distance geometry and applications. Spec Issue Discrete Appl Math 197:1–144
Nilges M, O’Donoghue SI (1998) Ambiguous NOEs and automated NOE assignment. Prog Nucl Magn Reson Spectrosc 32(2):107–139
Patterson AL (1944) Ambiguities in the X-ray analysis of crystal structures. Phys Rev 65:195–201
Rader AJ, Hespenheide BM, Kuhn LA, Thorpe MF (2002) Protein unfolding: rigidity lost. PNAS 99:3540–3545
Saxe J (1979) Embeddability of weighted graphs in k-space is strongly NP-hard. In: Conference in communications control and computing, pp 480–489
Schneider MN, Seibald M, Lagally P, Oeckler O (2010) Ambiguities in the structure determination of antimony tellurides arising from almost homometric structure models and stacking disorder. J Appl Cryst 43:1011–1020
Senechal M (2008) A point set puzzle revisited. Eur J Comb 29:1933–1944
Sivia DS (2011) Elementary scattering theory. Oxford University Press, Oxford
Skiena S, Smith W, Lemke P (1990) Reconstructing sets from interpoint distances. In Sixth ACM symposium on computational geometry, pp 332–339
Tay TS (1984) Rigidity of multi-graphs I: linking rigid bodies in n-space. J Comb Theory Ser B 36:95–112
Thorpe MF, Duxbury PM (eds) (1999) Rigidity theory and applications. Kluwer Academic, Dordrecht
Tucker MG, Keen DA, Dove MT, Goodwin AL, Huie Q (2007) RMCProfile: reverse Monte Carlo for polycrystalline materialss. J Phys Condens Matter 19:335218
Voller Z, Wu Z (2013) Distance geometry methods for protein structure determination, pp 139–159. In Mucherino et al. (2013)
Whiteley W (2005) Counting out to the flexibility of molecules. Phys Biol 2:S116–S126
Wu D, Wu Z (2007) An updated geometric build-up algorithm for solving the molecular distance geometry problems with sparse data. J Glob Optim 37:661–672
Wuthrich K (1989) The development of nuclear magnetic resonance spectroscopy as a technique for protein structure determination. Acc Chem Res 22(1):36–44
Acknowledgments
Support for work at Michigan State University by the MSU foundation is gratefully acknowledged. Collaborations with Pavol Juhas, Luke Granlund, Saurabh Gujarathi, Chris Farrow and Connor Glosser are much appreciated. PMD, CL and AM would like to thank Leo Liberti for interesting and motivating discussions. AM was supported by a grant of University of Rennes 1 for the development of international collaborations. PMD and CL were financially supported by the Brazilian research agencies FAPESP and CNPq. Work in the Billinge group was supported by the US National Science foundation DMREF program through grant: DMR-1534910.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Billinge, S.J.L., Duxbury, P.M., Gonçalves, D.S. et al. Assigned and unassigned distance geometry: applications to biological molecules and nanostructures. 4OR-Q J Oper Res 14, 337–376 (2016). https://doi.org/10.1007/s10288-016-0314-2
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10288-016-0314-2