Abstract
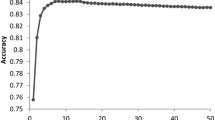
Peptide-MHC binding is an important prerequisite event and has immediate consequences to immune response. Those peptides binding to MHC molecules can activate the T-cell immunity, and they are useful for understanding the immune mechanism and developing vaccines for diseases. Accurate prediction of the binding between peptides and MHC-II molecules has long been a challenge in bioinformatics. Recently, instead of differentiating peptides as binder or non-binder, researchers are more interested in making predictions directly on peptide binding affinities. In this paper, we investigate the use of relevance vector machine to quantitatively predict the binding affinities between MHC-II molecules and peptides. In our scheme, a new encoding scheme is used to generate the input vectors, and then by using relevance vector machine we develop the prediction models on the basis of binding cores, which are recognized in an iterative self-consistent way. When applied to three MHC-II molecules DRB1*0101, DRB1*0401 and DRB1*1501, our method produces consistently better performance than several popular quantitative methods, in terms of cross-validated squared error, cross-validated correlation coefficient, and area under ROC curve. All evidences indicate that our method is an effective tool for MHC-II binding affinity prediction.
Similar content being viewed by others
References
Donnes P, Elofsson A (2002) Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinform 3:25
Rammennsee HG, Friede T, Stevanovic S (1995) MHC ligands and peptide motifs: first listing. Immunogenetics 41:178–228
Brusic V, Rudy G, Honeyman G et al. (1998) Prediction of MHC class II-binding peptides using an evolutionary algorithm and artificial neural network. Bioinformatics 14(2):121–130
Nielsen M, Lundegaard C, Worning P et al. (2004) Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics 20(9):1388–1397
Karpenko O, Shi J, Dai Y (2005) Prediction of MHC class II binders using the ant colony search strategy. Artif Intell Med 35(1–2):147–156
Murugan N, Dai Y (2005) Prediction of MHC class II binding peptides based on an iterative learning model. Immunome Res 1:6
Cui J, Han LY, Lin HH et al. (2007) Prediction of MHC-binding peptides of flexible lengths from sequence-derived structural and physicochemical attributes. Mol Immunol 44(5):866–877
Salomon J, Flower DR (2006) Predicting Class II MHC-Peptide binding: a kernel based approach using similarity scores. BMC Bioinform 7:501
Doytchinova IA, Flower DR (2003) Towards the in silico identification of class II restricted T-cell epitopes: a partial least squares iterative self-consistent algorithm for affinity prediction. Bioinformatics 19(17):2263–2270
Wan J, Liu W, Xu Q et al. (2006) SVRMHC prediction server for MHC-binding peptides. BMC Bioinform 7:463
Tipping ME (2000) The Relevance Vector Machine. In: Solla SA, Leen TK, Müller K-R (eds) Advances in neural information processing systems, vol 12. MIT Press, Cambridge, pp 652–658
Maglogiannis I, Zafiropoulos E, Anagnostopoulos I (2007) An intelligent system for automated breast cancer diagnosis and prognosis using SVM based classifiers. Appl Intell. doi:10.1007/s10489-007-0073-z
Furey TS, Cristianini N, Duffy N et al. (2000) Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 16(10):906–914
Rogers S, Girolami M (2005) A Bayesian regression approach to the inference of regulatory networks from gene expression data. Bioinformatics 21(14):3131–3137
Wei L, Yang Y, Nishikawa RM, Wernick MN, Edwards A (2005) Relevance vector machine for automatic detection of clustered microcalcifications. IEEE Trans Med Imaging 24(10):1278–1285
Li Y, Campbell C, Tipping M (2002) Bayesian automatic relevance determination algorithms for classifying gene expression data. Bioinformatics 18(10):1332–1339
Mika S, Rost B (2003) UniqueProt: creating representative protein-sequence sets. Nucl Acids Res 31(13):3789–3791
Chang S, Ghosh D, Kirschner D et al. (2006) Peptide length-based prediction of peptide-MHC class II binding. Bioinformatics 22(22):2761–2767
Liu W, Meng X, Xu Q et al. (2006) Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models. BMC Bioinform 7:182
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, W., Liu, J. & Niu, Y. Quantitative prediction of MHC-II peptide binding affinity using relevance vector machine. Appl Intell 31, 180–187 (2009). https://doi.org/10.1007/s10489-008-0121-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-008-0121-3