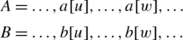Abstract
Pattern matching with wildcards is a challenging topic in many domains, such as bioinformatics and information retrieval. This paper focuses on the problem with gap-length constraints and the one-off condition (The one-off condition means that each character can be used at most once in all occurrences of a pattern in the sequence). It is difficult to achieve the optimal solution. We propose a graph structure WON-Net (WON-Net is a graph structure. It stands for a network with the weighted centralization measure based on each node’s centrality-degree. Its details are given in Definition 4.1) to obtain all candidate matching solutions and then design the WOW (WOW stands for pattern matching with wildcards based on WON-Net) algorithm with the weighted centralization measure based on nodes’ centrality-degrees. We also propose an adjustment mechanism to balance the optimal solutions and the running time. We also define a new variant of WOW as WOW-δ. Theoretical analysis and experiments demonstrate that WOW and WOW-δ are more effective than their peers. Besides, the algorithms demonstrate an advantage on running time by parallel processing.


















Similar content being viewed by others
References
Pisanti N, Crochemore M, Grossi R, Sagot M-F (2005) Bases of motifs for generating repeated patterns with wild cards. IEEE/ACM Trans Comput Biol Bioinform 2:40–50
On B-W, Lee I (2011) Meta similarity. Appl Intell 35(3):359–374
Xiao L, Wissmann D, Brown M, Jablonski S (2004) Information extraction from the web: system and techniques. Appl Intell 21(2):195–224
Bille P, Gørtz IL, Vildhøj HW, Wind DK (2010) String matching with variable length gaps. In: String processing and information retrieval—17th international symposium, vol 6393, pp 385–394
Zhou B, Pei J (2012) Aggregate keyword search on large relational databases. Knowl Inf Syst 30(2):283–318
Hofmann K, Bucher P, Falquet L, Bairoch A (1999) The PROSITE database, its status in 1999. Nucleic Acids Res 27:215–219
Bucher P, Bairoch A (1994) A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. In: Proceedings of the 2nd international conference on intelligent systems for molecular biology, pp 53–61
Navarro G, Raffinot M (2002) Flexible pattern matching in strings—practical on-line search algorithms for texts and biological sequences. Cambridge University Press, Cambridge
Cole R, Gottlieb L-A, Lewenstein M (2004) Dictionary matching and indexing with errors and don’t cares. In: Proceedings of the 36th ACM symposium on the theory of computing. ACM, New York, pp 91–100
Ménard PA, Ratté S (2011) Classifier-based acronym extraction for business documents. Knowl Inf Syst 29(2):305–334
Sánchez D, Isern D (2011) Automatic extraction of acronym definitions from the web. Appl Intell 34(2):311–327
Ahmed CF, Tanbeer SK, Jeong B-S, Lee Y-K (2011) HUC-prune: an efficient candidate pruning technique to mine high utility patterns. Appl Intell 34(2):181–198
Shie B-E, Yu PS, Tseng VS (2012) Mining interesting user behavior patterns in mobile commerce environments. Appl Intell. doi:10.1007/s10489-012-0379-3
Agrawal R, Srikant R (1995) Mining sequential patterns. In: Proc of ICDE, Taipei, pp 3–14
Chen G, Wu X, Zhu X, Arslan AN, He Y (2006) Efficient string matching with wildcards and length constraints. Knowl Inf Syst 10(4):399–419
Fischer MJ, Paterson MS (1974) String matching and other products. Technical report, Massachusetts Institute of Technology, Cambridge, MA, USA
Zhang M, Kao B, Cheung DW, Yip KY (2005) Mining periodic patterns with gap requirement from sequences. In: Proceedings of ACM SIGMOD, Baltimore, Maryland, USA, pp 623–633
Ding B, Lo D, Han J, Khoo S (2009) Efficient mining of closed repetitive gapped subsequences from a sequence database. In: Proceedings of IEEE 25th international conference on data engineering (ICDE 09), Shanghai, PR China, 2009. IEEE Comput Soc, Los Alamitos, pp 1024–1035
Min F, Wu X, Lu Z (2009) Pattern matching with independent wildcard gaps. In: Eighth IEEE international conference on dependable, autonomic and secure computing (DASC-2009), Chengdu, China, pp 194–199
Guo D, Hong X, Hu X, Gao J, Liu Y, Wu G, Wu X (2011) A bit-parallel algorithm for sequential pattern matching with wildcards. Cybern Syst 42(6):382–401
Wang H, Xie F, Hu X, Li P, Wu X (2010) Pattern matching with flexible wildcards and recurring characters. In: 2010 IEEE international conference on granular computing (GrC 2010), Silicon Valley, USA, 2010. IEEE Comput Soc, Los Alamitos, pp 782–786
Wu Y, Wu X, Jiang H, Min F (2011) A heuristic algorithm for MPMGOOC. Chin J Comput 32(8):1452–1462
Chang Y-I, Chen J-R, Hsu M-T (2010) A hash trie filter method for approximate string matching in genomic databases. Appl Intell 33(1):21–38
He D, Wu X, Zhu X (2007) SAIL-APPROX: an efficient on-line algorithm for approximate pattern matching with wildcards and length constraints. In: Proceedings of the IEEE international conference on bioinformatics and biomedicine (BIBM’07), Silicon Valley, USA, pp 151–158
Dorneles CF, Gonçalves R, Mello RS (2011) Approximate data instance matching: a survey. Knowl Inf Syst 27(1):1–21
Goethals B, Laurent D, Page WL, Dieng CT (2012) Mining frequent conjunctive queries in relational databases through dependency discovery. Knowl Inf Syst. doi:10.1007/s10115-012-0526-5
Xiong N, Funk P (2008) Concise case indexing of time series in health care by means of key sequence discovery. Appl Intell 28(3):247–260
Xie F, Wu X, Hu X, Gao J, Guo D, Fei Y, Hua E (2011) MAIL: mining sequential patterns with wildcards. Int J Data Min Bioinforma. http://www.inderscience.com/coming.php?ji=189&jc=ijdmb&np=9&jn=International%20Journal%20of%20Data%20Mining%20and%20Bioinformatics
Martínez-Trinidad JF, Carrasco-Ochoa JA, Ruiz-Shulcloper J (2011) RP-miner: a relaxed prune algorithm for frequent similar pattern mining. Knowl Inf Syst 27(3):451–471
Wu Y, Wu X, Min F, Li Y (2010) A nettree for pattern matching with flexible wildcard constraints. In: Proceedings of the 2010 IEEE international conference on information reuse and integration (IRI 2010), Las Vegas, USA, pp 109–114
Liu Y, Wu X, Hu X, Gao J et al (2009) Pattern matching with wildcards based on key character location. In: Proceedings of the 2009 IEEE international conference on information reuse and integration (IRI 2009), Las Vegas, USA, pp 167–170
National Center for Biotechnology Information (2009) GenBank sequences from pandemic (H1N1) 2009 viruses. http://www.ncbi.nlm.nih.gov/genomes/FLU/SwineFlu.html
Artificial data. http://dmic.hfut.edu.cn/HFUT_DMIC/DanGuo/test
Acknowledgements
This work is supported by the National High Technology Research and Development Program of China (863 Program) under grant 2012AA011005, the National 973 Program of China under grant 2013CB329604, the National Natural Science Foundation of China (NSFC) under grant 61229301, the China Postdoctoral Science Foundation under grant 2012M511403, and the US National Science Foundation (NSF) under grant CCF-0905337.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
1.1 A.1 Proof of Formula 5.2
Proof
\(D_{node_{dep} \in S_{i,g}}\) is the number of paths from root nodes to leaf nodes through node dep in S i,g . It is equivalent to |S i,g | (the number of occurrences in S i,g ). Similarly, \(D_{node_{dep} \in S_{j,g}} = |S_{j,g}|\).
According to Definition 5.1, \(d_{\mathit{occ}_{k,i,g},S_{j,g}} =\allowbreak \sum_{node \in \mathit{occ}_{{k,i,g}}} D_{node \in S_{j,g}}\). Therefore, LWC[occ h,i,g ] in Formula 5.2 holds.

Therefore, CWC[S i,g ] in Formula 5.2 is confirmed. □
1.2 A.2 Proof of Theorems in Sect. 6.2
Theorem 6.1
If WON-Net(P,S) is null, WOW is complete. There exists \(|O_{\mathit{PMG}}(P,S)|= |O_{\mathit{PMGO}}(P,S)|=|O_{\mathit{PMGO\_MAX}}(P,S)|=0\).
Proof
If WON-Net(P,S) is null, it means that there does not exist P in S. □
Property 6.1
\(|O_{\mathit{PMGO\_Tg}}(P,S)|\leq \min\{ \min_{0 \le j < m}\{ C_{T_{g},j}\} ,\allowbreak \lfloor\frac{s_{T_{g},\max} - s_{T_{g},\min} + 1}{G_{N}} \rfloor\}\), where \(O_{\mathit{PMGO\_Tg}}(P,S)\) is a subset of O PMGO (P,S) on T g , \(C_{T_{g},j}\) is the number of nodes on the j-th layer in T g , \(s_{T_{g},\max}\) and \(s_{T_{g},\min}\) are respectively the maximal and minimal node number in T g , and G N is the minimal global length of P.
Explanation
For each \(T_{g}, |O_{\mathit{PMGO\_Tg}}(P,S)|\) is no more than \(\min_{0 \le j < m}\{ C_{T_{g},j}\}\) based on the graph structure T g , and no more than \(\lfloor\frac{s_{T_{g},\max} - s_{T_{g},\min} + 1}{G_{N}} \rfloor\) with the global length constraint under the one-off condition. As in T 1 and T 2 in Example 3, \(|O_{\mathit{PMGO\_T1}}(P,S)| =\min\{3, \lfloor\frac{20 - 12 + 1}{4} \rfloor\}=\{3,2\}=2\), and \(|O_{\mathit{PMGO\_T2}}(P,S)| =\min\{1,\lfloor\frac{31 - 24 + 1}{4} \rfloor\}=\{1,2\}=1\).
Theorem 6.2
If

WOW is complete.
Proof
\(|O_{\mathit{PMGO}}(P,S)| = \sum_{0 \le g < |\mathit{Net}|} |O_{\mathit{PMGO\_Tg}}(P,S)| \le\sum_{0 \le g < |\mathit{Net}|} \min\{ \min_{0 \le j < m}\{ C_{T_{g},j}\} , \lfloor\frac{s_{T_{g},\max} -s_{T_{g},\min} + 1}{G_{N}} \rfloor\}\). WOW is complete based on Property 6.1. □
Theorem 6.3
If P does not have duplicate sub-patterns (i.e. no p i repeats (0≤i≤m−1)), WOW is complete.
Theorem 6.4
If each gap has the same upper and lower limits (∃N i =M j for ∀i, 0≤i<m−1), WOW is complete.
Proof
LMO/RMO in WOW is a transform of SAIL/RSAIL based on graph structure WON-Net(P,S) respectively. According to Theorems 2 and 4 in our previous work [16], we know that SAIL/RSAIL is complete while P does not have duplicate sub-patterns or each gap has the same upper and lower limits. Therefore, Theorems 6.3 and 6.4 are proved. □
Definition 6.1
If a node appears on different layers in WON-Net, the node is a free-node. In Fig. 1, there are six free-nodes: 4, 5, 6, 15, 16 and 17.
Theorem 6.5
If WON-Net(P,S) does not have free-nodes, WOW is complete.
Proof
There is the inverse negation of Theorem 6.5: “If WOW is incomplete, WON-Net(P,S) has free-nodes”.
Let \(O_{\mathit{PMGO\_LMO}}(P,S)\) be the occurrence set by the LMO strategy in WOW, and \(O_{\mathit{PMGO\_MAX}}(P,S)\) be the optimal one. Suppose \(|O_{\mathit{PMGO\_LMO}}(P,S)|=1\) and \(|O_{\mathit{PMGO\_MAX}}(P,S)|=2\). Let occurrences A=〈a[0],a[1],…,a[i],…,a[m−1]〉, \(B=\langle b[0],b[1],\ldots,b[i],\ldots,\allowbreak b[m-1]\rangle \in O_{\mathit{PMGO\_MAX}}(P,S)\), and \(\textit{Occ}=\langle \textit{occ}[0],\allowbreak \textit{occ}[1],\ldots, \textit{occ}[i],\ldots, \textit{occ}[m-1]\rangle \in O_{\mathit{PMGO\_LMO}}(P,S)\). Occ and A, Occ and B do not satisfy the one-off condition. There exist a[u]=occ[i] and b[w]=occ[k] (i≠k, 0≤u,w,i,k<m).
If u=w, there exists a[u]=b[w]. A and B do not satisfy the one-off condition. It is in contradiction with the assumption that A, \(B\in O_{\mathit{PMGO\_MAX}}(P,S)\).
Let u<w. There are two possible cases as follows.
-
1.
u≠i or w≠k
If u≠i and a[u]=occ[i], the indexed position number a[u] is a free-node. It appears both on the u-th layer and the i-th layer in WON-Net(P,S). WON-Net(P,S) has free-nodes. It is proved while u≠i. Similarly, it can also be proved while w≠k.
-
2.
u=i and w=k
There is a[u]=occ[u]<b[u]<b[w]=occ[w]<a[w] according to the LMO strategy.
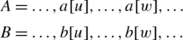
There must exist a[t]<b[t] and b[t+1]<a[t+1], u≤t≤w−1.

Otherwise, there are two possible cases:
-
a[t]≥b[t] and a[t+1]>b[t+1] (u≤t≤w−1). It is in contradiction with a[u]<b[u].
-
a[t]<b[t] and a[t+1]≤b[t+1] (u≤t≤w−1). It is in contradiction with b[w]<a[w].
So, there exist a[t]<b[t] and b[t+1]<a[t+1] (u≤t≤w−1).
Thus there exists N t ≤b[t+1]−b[t]<a[t+1]−b[t]<a[t+1]−a[t]≤M t (u≤t≤w−1).
Under the gap-length constraints, 〈b 0,b 1,…,b u ,…,b t ,a t+1,…,a w ,…,a m−1〉 is an occurrence of P in S. Thus, LMO will obtain another occurrence 〈b 0,b 1,…,b u ,…,b t ,a t+1,…,a w ,…,a m−1〉. \(O_{\mathit{PMGO\_LMO}}(P,S)= \{\textit{Occ},\langle b_{0},\allowbreak b_{1},\ldots, b_{u},\ldots, b_{t},a_{t+1},\ldots, a_{w},\ldots, a_{m-1}\rangle \}\). It is in contradiction with the assumption that \(|O_{\mathit{PMGO\_LMO}}(P,S)|\allowbreak =1<\allowbreak |O_{\mathit{PMGO\_MAX}}(P,S)|=2\).
Therefore, the inverse negation that if WOW is incomplete, WON-Net(P,S) has free-nodes is true, and Theorem 6.5 is proved.
In addition, there also exists the completeness of O PMG (P,S) based on WON-Net(P,S) as follows. □
Theorem 6.6
\(|O_{\mathit{PMG}}(P,S)|=\sum_{0 \le g < |\mathit{Net}|} D_{T_{g}}\) and \(D_{T_{g}} = D_{T_{g},0} =\cdots = D_{T_{g},j} =\cdots= D_{T_{g},m - 1}\), where \(D_{T_{g},j} = \sum_{\{ node_{g,j}\}} D(\mathit{node}_{g,j}) |T_{g}\), and \(D_{T_{g},j}\) is the sum of nodes’ centrality-degrees on the j-th layer in T g .
Proof
O PMG (P,S) is a set which contains all occurrences of P in S. O PMG (P,S) can be divided into the sum of \(O_{\mathit{PMG\_Tg}}(P,S)\) based on Property 4.1(2). \(D_{T_{g},0}\) is the sum of the root nodes’ centrality-degrees in T g . \(D_{T_{g},0}\) is equivalent to \(O_{\mathit{PMG\_Tg}}(P,S), 0\leq g<|\mathit{Net}|, 0\leq j<m\).

where \(n^{k}_{g,j}\) is k-th node on the j-th layer in T g , and \(C_{T_{g},j}\) is the number of nodes on the j-th layer in T g . □
1.3 A.3 The random data generator in Sect. 7.2
Artificial data are generated by the data generator to test the completeness of the pattern matching algorithms under the one-off condition. The data generator is described in [23]. The data generator is based on Algorithm DG (Algorithm 6) as follows. It can be proved with the complete solution of P in S under the one-off condition.
Given a character alphabet Σ, the gap-length constraints of pattern P, the length of the subject sequence S, and the maximal support max_sup, the data generator can produce S with the exact support value of P in S under the one-off condition. The support is equivalent to |OPMG(P,S)| and no more than max_sup.
The artificial data and the data generator program can be downloaded from the following website: http://dmic.hfut.edu.cn/HFUT_DMIC/DanGuo/test.
Rights and permissions
About this article
Cite this article
Guo, D., Hu, X., Xie, F. et al. Pattern matching with wildcards and gap-length constraints based on a centrality-degree graph. Appl Intell 39, 57–74 (2013). https://doi.org/10.1007/s10489-012-0394-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-012-0394-4