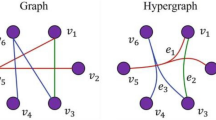
Abstract
Link prediction addresses the problem of finding potential links that may form in the future. Existing state of art techniques exploit network topology for computing probability of future link formation. We are interested in using Graphical models for link prediction. Graphical models use higher order topological information underlying a graph for computing Co-occurrence probability of the nodes pertaining to missing links. Time information associated with the links plays a major role in future link formation. There have been a few measures like Time-score, Link-score and T_Flow, which utilize temporal information for link prediction. In this work, Time-score is innovatively incorporated into the graphical model framework, yielding a novel measure called Temporal Co-occurrence Probability (TCOP) for link prediction. The new measure is evaluated on four standard benchmark data sets : DBLP, Condmat, HiePh-collab and HiePh-cite network. In the case of DBLP network, TCOP improves AUROC by 12 % over neighborhood based measures and 5 % over existing temporal measures. Further, when combined in a supervised framework, TCOP gives 93 % accuracy. In the case of three other networks, TCOP achieves a significant improvement of 5 % on an average over existing temporal measures and an average of 9 % improvement over neighborhood based measures. We suggest an extension to link prediction problem called Long-term link prediction, and carry out a preliminary investigation. We find TCOP proves to be effective for long-term link prediction.


Similar content being viewed by others
References
Adamic LA, Adar E (2001) Friends and neighbors on the web. Soc Networks 25:211–230
Aggarwal CC, Xie Y, Philip S (2014) A framework for dynamic link prediction in heterogeneous networks. Statistical Analysis and Data Mining 7(1):14–33
Calders T, Goethals B (2002) Mining all non-derivable frequent itemsets. In: Proceedings of the 6th European Conference on Principles of Data Mining and Knowledge Discovery, PKDD’02. Springer-Verlag, pp 74–85
Choudhary P, Mishra N, Sharma S, Patel R (2013) Link score: A novel method for time aware link prediction in social network. ICDMW
da Silva Soares PR, Prudêncio RBC (2012) Time series based link prediction. In: The 2012 International Joint Conference on Neural Networks (IJCNN). IEEE, pp 1–7
Davis DA, Lichtenwalter R, Chawla NV (2013) Supervised methods for multi-relational link prediction. Soc Netw Anal Min 3(2):127–141
Davis J, Goadrich M (2006) The relationship between precision-recall and roc curves. In: Proceedings of the 23rd international conference on Machine learning, ICML ’06. ACM, pp 233–240
Dunlavy DM, Kolda TG, Acar E (2011) Temporal link prediction using matrix and tensor factorizations. ACM Trans Knowl Discov Data (TKDD) 5(2):10
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The weka data mining software: an update. SIGKDD Explorations Newsletter 11(1):10–18
Hasan MA, Chaoji V, Salem S, Zaki M (2006) Link prediction using supervised learning. In: Proceedings of SDM 06 workshop on Link Analysis Counterterrorism and Security
Lakshmi TJ, Bhavani SD (2015) Enhancement to community-based multi-relational link prediction using co-occurrence probability feature. In: Proceedings of the Second ACM IKDD Conference on Data Sciences, CoDS ’15. ACM, pp 86–91
Jelinek F (1977) Continuous speech recognition. SIGART Bulletin 61:33–34
Katz L (1953) A new status index derived from sociometric analysis. Psychometrika 18(1):39–43
Kindermann R (1980) Markov random fields and their applications, Providence, R.I
Lakshmi TJ, Bhavani SD (2014) Heterogeneous link prediction based on multi relational community information. In: 6th International Conference on Communication Systems and Networks, COMSNETS 2014, pp 1–4
Liben-Nowell D, Kleinberg J (2007) The link-prediction problem for social networks. Journal of The American Society For Information Science and Technology 58(7):1019–1031
Lichtenwalter R, Chawla NV (2012) Link prediction: fair and effective evaluation. In: ASONAM. IEEE Computer Society, pp 376–383
Lichtenwalter R, Chawla NV (2012) Vertex collocation profiles: subgraph counting for link analysis and prediction. In: WWW. ACM, pp 1019–1028
Lichtenwalter RN, Chawla NV (2011) Lpmade: Link prediction made easy. J Mach Learn Res 12:2489–2492
Lichtenwalter RN, Lussier JT, Chawla NV (2010) New perspectives and methods in link prediction. In: Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD’10. ACM, pp 243–252
Mooij JM (2010) libDAI: A free and open source C++ library for discrete approximate inference in graphical models. J Mach Learn Res 11:2169–2173
Munasinghe L (2013) Time-aware methods for link prediction in social networks. PhD THesis, The Graduate University for Advanced Studies
Munasinghe L, Ichise R (2011) Time aware index for link prediction in social networks. In: DaWaK, volume 6862 of Lecture Notes in Computer Science. Springer, pp 42–353
Newman MEJ (2001) Clustering and preferential attachment in growing networks. Phys Rev E 64(2):025102
Pavlov D, Mannila H, Smyth P (2000) Probabilistic models for query approximation with large sparse binary data sets. In: UAI-2000. Morgan Kaufmann Publishers, pp 465–472
Raghavan V, Bollmann P, Gwang JS (1989) A critical investigation of recall and precision as measures of retrieval system performance, vol 7. ACM, pp 205–229
Spiegelhalter DJ, Lauritzen SL (1988) Local computations with probabilities on graphical structures and their application to expert systems. J R Stat Soc Ser B Methodol 50(2):157– 224
Sun Y, Barber R, Gupta M, Aggarwal CC, Han J (2011) Co-author relationship prediction in heterogeneous bibliographic networks. In: Proceedings of the International Conference on Advances in Social Networks Analysis and Mining, ASONAM ’11. IEEE Computer Society, p 2011
Sun Y, Han J, Aggarwal CC, Chawla NV (2012) When will it happen?: relationship prediction in heterogeneous information networks. In: Proceedings of the 5th ACM international conference on Web search and data mining. ACM, pp 663–672
Tylenda T, Angelova R, Bedathur S (2009) Towards time-aware link prediction in evolving social networks. In: Proceedings of the 3rd Workshop on Social Network Mining and Analysis, SNA-KDD ’09. ACM, pp 1–10
Wang C, Satuluri V, Parthasarathy S (2007) Local probabilistic models for link prediction. In: Proceedings of 7th IEEE International Conference on Data Mining, ICDM ’07. IEEE Computer Society, pp 322–331
Wang C, Han J, Jia Y, Tang J, Zhang D, Yintao Y, Guo J (2010) Mining advisor-advisee relationships from research publication networks. In: Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’10. ACM, pp 203–212
Yang Y, Chawla NV, Sun Y, Han J (2012) Predicting links in multi-relational and heterogeneous networks. In: 12th IEEE International Conference on Data Mining, ICDM 2012, Brussels, Belgium, December 10-13, 2012, pp 755–764
Acknowledgments
Partial support from UGC UPE-II, University of Hyderabad is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A: Graphical model framework
The notation of undirected graphical model called Markov Random Field [14] is given in this section.
Notation Given a graph G=(V,E) where random variables(X) are indexed by V, i.e X={X v },v∈V is probabilistic model, if it satisfies the following properties:
-
1.
Pair wise Markov Property : Any two non adjacent variables are conditionally independent.
$$ X_{u}~ \perp\!\!\!\perp ~X_{v}, if (u,v) \notin E $$(3) -
2.
Local Markov Property : A variable is conditionally independent of all the other variables given its neighbors:
$$ X_{u}~ \perp\!\!\!\perp ~X_{V-\Gamma(v)} ~|~\Gamma(v) $$(4)where Γ(v) is set of neighbors of v.
-
3.
Global Markov Property : Two sets A,B⊆V are conditionally independent, given their separator set S = A∩B in G.
$$ X_{A} \perp\!\!\!\perp X_{B}~|~X_{S} $$(5)
Whenever a general graph exhibits the Markovian properties, it can be factorized over cliques of G.i.e
where ϕ c (X c ) is clique potential of max Clique C. If G is directed, this graphical model is called Bayesian Network and if it is undirected, it is called Markov Random Field(MRF).
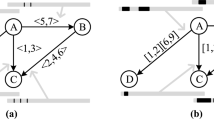
Appendix B: Example illustration of TCOP on snapshot of DBLP dataset
The joint probability inference of the nodes 195 and 6587 using belief propagation in junction tree is illustrated here.
The junction tree of Fig. 1b is shown in Fig. 3 and the corresponding clique potential tables computed using Algorithm 1 are shown in Tables 9, 10 and 11.
Junction tree of Fig 1b
We can see in Fig. 3 that there are 3 cliques and 2 separators.
Cliques :
Separators:
There will be one clique potential corresponding to each clique. In the example, the potentials are \(\phi _{C_{1}}, \phi _{C_{2}}~ and~ \phi _{C_{3}}\). These potential Tables are shown in Tables 9, 10 and 11.
The belief propagation in junction tree has two steps: Upward belief propagation and downward belief propagation.
1.1 Belief propagation in upward direction
-
For each leaf in the junction tree, send a message to its parent. The message is the marginal of its table, summing out any variable not in the separator.
$$ for~x \in S(C,C^{\prime}),~ \phi_{S(C,C^{\prime})}(x) = \sum\limits_{x\in C^{\prime}, x \notin S}P(x) $$(7)That means, every leaf clique node C ′ sends the potential table computed by (7), to its parent clique, separated by separator S(C,C ′).
In the above example illustration, Message from C 2→C 1 : Marginalize \(\phi _{C_{2}}\) to S(C 1,C 2) and is shown in Table 12. In the same way, message from C 3→C 1 can be obtained by marginalizing \(\phi _{C_{3}}\) to S(C 1,C 3), i.e,6587, 37227, as shown in Table 13.
-
When a parent Clique(say C) receives a message from a child (say C ′), it multiplies its table by the message table to obtain its new table and thus updates for all child nodes C ′.
$$ \phi_{C}(x)= \prod\limits_{C^{\prime}} \phi_{C}(x) \phi_{S(C,C^{\prime})}(x) $$(8)Continuing the same example as above,
Update C 1 by the message from its child C 2 to get Table 14.
Update C 1 by the message from its child C 3 to get Table 15.
-
When a parent receives messages from all its children, it repeats the process. This process continues until the root receives messages from all its children. In the above example, the tree is of height 1. So, therefore, the process stops in one iteration. The final potential of clique C 1 will be as in Table 16
After updations to clique potentials are carried out in upward pass, the downward pass is initiated.
1.2 Belief propagation in downward direction
This step reverses upward pass, starting at the root.
-
The root(say C) sends a message to each of its children. More specifically, the root divides its current table by the message received from the child through the separator(say S(C,C ′)), marginalizes the resulting table to the separator, and sends the result to the child.
$$ \phi_(x)= \sum\limits_{x \in C, x \notin S(C,C^{\prime})}\frac{\phi_{C^{\prime}}(x)}{\phi_{S(C,C^{\prime})}(x)} $$(9)
In the example, the message from clique C 1→C 2 is computed, first by dividing C 1 by S(C 1,C 2) as shown in Table 17 and then marginalizing it to S(C 1,C 2) as shown in Table 18.
In the same way, the message from clique C 1→C 3 is computed, first by dividing C 1 by S(C 1,C 3) as shown in Table 19 and then marginalizing it to S(C 1,C 3) as shown in Table 20.
-
Each child(C ′) multiplies its table by its parent’s(C) table and repeats the process (acts as a root) until leaves are reached.
$$ \phi_{C^{\prime}}(x)=\prod\phi_{C}(x)\phi_{S}(x) $$(10)Thus, final potential table of clique is obtained by multiplying the table of C 2 by message obtained by its parent C 1 through its separator S(C 1,C 2), which is shown in table . Similar is the case for getting final potential of clique C 3 (Tables 21 and 22).
Thus, potential of each clique is calculated as a joint marginal of its variables, by marginalizing(sum out) over the variables that are not needed. Junction Tree Algorithm finishes the upward and downward passes, Potentials will be equal to Marginals. To infer the joint probability of two variables, we will pick those two variables from any cliques, and multiply their potentials.
For instance, to compute the joint probability of nodes 195 and 6587, first pick the clique containing 195, i.e, C 2 and marginalize the two entries corresponding to the value 1 of node 195, which is 0.2158, and then pick the clique containing node 6587, i.e, C 3 and marginalize the entries corresponding to value 1 for node 6587, which is 0.01438. The joint probability is simply the product of 0.2158 * 0.01438 = 0.003103204.
Rights and permissions
About this article
Cite this article
Jaya Lakshmi, T., Durga Bhavani, S. Temporal probabilistic measure for link prediction in collaborative networks. Appl Intell 47, 83–95 (2017). https://doi.org/10.1007/s10489-016-0883-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-016-0883-y