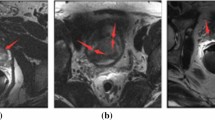
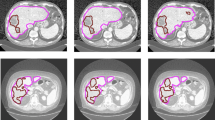
Abstract
The low resolution of thick CT/MR with large spacing will lead to misdiagnosis and bring a huge difficulty to the automatic organ or lesion segmentation. Especially in automatic pancreatic tumor segmentation, due to the limitation of equipment, the tumors only exist in a few slices, and the continuity between slices of 3D CT/MR image is poor. Besides, the features of tumors, such as the size, shape, location, intensity and so on, vary dramatically according to different cases. The fuzzy boundaries of small tumors also bring great uncertainty to the segmentation task. Aiming to solve those problems, we introduce the LLE-based interpolation neural network into the pancreatic tumor segmentation task, which mainly includes the following improvements: 1) We utilize local linear embedding (LLE) to model the relationship between adjacent slices and the interpolated slice. It adapts the spatial transformation of the organ between slices. 2) Neural network, combining with the LLE module, is designed to significantly enhance the image resolution, thus can generate more continuous and clearer images for each sequence. 3) Multiscale cascade strategy is adopted in the network to reduce the influence of drastic changes in tumor size on segmentation results. Experiments are carried out in MR images with 3.5mm thickness provided by Changhai hospital, and CT images on Medical Segmentation Decathlon pancreatic tumor challenge, respectively. The results show that our proposed model has a sensitivity of 96.55% and a dice coefficient of 62.8% (± 25.8%) on the MR dataset, and a dice coefficient of 50.6% (± 30.9%) on the CT dataset.








Similar content being viewed by others
References
Chen MY, Woodruff MA, Kua B, Rukin NJ (2021) Rapid segmentation of renal tumours to calculate volume using 3d interpolation. Journal of Digital Imaging. https://doi.org/10.1007/s10278-020-00416-z
El-Hag NA, Sedik A, El-Banby GM, El-Shafai W, Khalaf AAM, Al-Nuaimy W, Abd El-Samie FE, El-Hoseny HM (2019) Utilization of image interpolation and fusion in brain tumor segmentation. International Journal for Numerical Methods in Biomedical Engineering. https://doi.org/10.1002/cnm.3449
Fang C, Li G, Pan C, Li Y, Yu Y (2019) Globally guided progressive fusion network for 3d pancreas segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 210–218
Gao Y, Beijbom O, Zhang N, Darrell T (2016) Compact bilinear pooling. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 317–326
Guo Y, Bi L, Ahn E, Feng D, Wang Q, Kim J (2020) A spatiotemporal volumetric interpolation network for 4d dynamic medical image. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 4726–4735
Guo Z, Zhang L, Lu L, Bagheri M, Summers RM, Sonka M, Yao J (2018) Deep logismos: Deep learning graph-based 3d segmentation of pancreatic tumors on ct scans. In: 2018 IEEE 15Th international symposium on biomedical imaging (ISBI 2018). IEEE, pp 1230–1233
Çiçek z, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3d u-net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 424–432
Isensee F, Jaeger PF, Kohl S, Petersen J, Maier-hein KH (2021) nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18(2):203–211
Isensee F, Petersen J, Klein A, Zimmerer D, Jaeger PF, Kohl S, Wasserthal J, Koehler G, Norajitra T, Wirkert S et al (2019) nnu-net: Self-adapting framework for u-net-based medical image segmentation. In: Bildverarbeitung für die medizin 2019. Springer, pp 22–22
Jia F, Liu J, Tai XC (2021) A regularized convolutional neural network for semantic image segmentation. Anal Appl 19(01):147–165
Jia H, Song Y, Huang H, Cai W, Xia Y (2019) Hd-net: Hybrid discriminative network for prostate segmentation in mr images. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 110–118
Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA (2018) H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes. IEEE Trans Med Imaging 37(12):2663–2674
Liang Y, Schott D, Zhang Y, Wang Z, Nasief H, Paulson E, Hall W, Knechtges P, Erickson B, Li XA (2020) Auto-segmentation of pancreatic tumor in multi-parametric mri using deep convolutional neural networks. Radiother Oncol 145:193–200
Liu S, Xu D, Zhou SK, Pauly O, Grbic S, Mertelmeier T, Wicklein J, Jerebko A, Cai W, Comaniciu D (2018) 3d anisotropic hybrid network: Transferring convolutional features from 2d images to 3d anisotropic volumes. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Springer International Publishing, pp 851–858
Liu S, Yuan X, Hu R, Liang S, Feng S, Ai Y, Zhang Y (2019) Automatic pancreas segmentation via coarse location and ensemble learning. IEEE Access 8:2906–2914
Maheshwari H, Goel V, Sethuraman R, Sheet D (2021) Distill DSM: Computationally efficient method for segmentation of medical imaging volumes. In: Medical Imaging with Deep Learning. https://openreview.net/forum?id=_n48l6YKc6d
Milletari F, Navab N, Ahmadi SA (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth international conference on 3d vision (3DV), pp 565–571. IEEE
Mo Y, Liu F, McIlwraith D, Yang G, Zhang J, He T, Guo Y (2018) The deep poincaré map: a novel approach for left ventricle segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 561–568
Nie D, Gao Y, Wang L, Shen D (2018) Asdnet: Attention based semi-supervised deep networks for medical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 370–378
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B (2018) Attention u-net: Learning where to look for the pancreas. arXiv:1804.03999
Peng C, Lin WA, Liao H, Chellappa R, Zhou SK (2020) Saint: spatially aware interpolation network for medical slice synthesis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 7750–7759
Sandrasegaran K, Lin Y, Asare-Sawiri M, Taiyini T, Tann M (2019) Ct texture analysis of pancreatic cancer. Eur Radiol 29(3):1067–1073
Shi G, Xiao L, Chen Y, Zhou SK Marginal loss and exclusion loss for partially supervised multi-organ segmentation 70:101,979. https://doi.org/10.1016/j.media.2021.101979. https://linkinghub.elsevier.com/retrieve/pii/S1361841521000256
Shi W, Caballero J, Huszár F, Totz J, Aitken AP, Bishop R, Rueckert D, Wang Z (2016) Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1874–1883
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70(1):7–30
Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B, Ronneberger O, Summers RM, Bilic P, Christ PF, Do RKG, Gollub M, Golia-Pernicka J, Heckers SH, Jarnagin WR, McHugo MK, Napel S, Vorontsov E, Maier-Hein L, Cardoso MJ (2019) A large annotated medical image dataset for the development and evaluation of segmentation algorithms
Taydaş O., Durhan G, Akpınar MG, Demirkazık FB (2019) Comparison of mri and us in tumor size evaluation of breast cancer patients receiving neoadjuvant chemotherapy. Eur J Breast Health 15 (2):119
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13(4):600–612
Wu Z, Wei J, Yuan W, Wang J, Tasdizen T (2020) Inter-slice image augmentation based on frame interpolation for boosting medical image segmentation accuracy. arXiv:2001.11698
Xia Y, Xie L, Liu F, Zhu Z, Fishman EK, Yuille AL (2018) Bridging the gap between 2d and 3d organ segmentation with volumetric fusion net. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Springer International Publishing, pp 445–453
Xia Y, Xie L, Liu F, Zhu Z, Fishman EK, Yuille AL (2018) Bridging the gap between 2d and 3d organ segmentation with volumetric fusion net. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 445–453
Yang D, Roth H, Xu Z, Milletari F, Zhang L, Xu D Searching learning strategy with reinforcement learning for 3d medical image segmentation Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, Yap PT, Khan A (eds), vol 11765, Springer International Publishing. https://doi.org/10.1007/978-3-030-32245-8_1. Decathlon
Yu Q, Shi Y, Sun J, Gao Y, Zhu J, Dai Y (2019) Crossbar-net: A novel convolutional neural network for kidney tumor segmentation in ct images. IEEE Transactions on Image Processing
Zhang D, Huang G, Zhang Q, Han J, Han J, Wang Y, Yu Y (2020) Exploring task structure for brain tumor segmentation from multi-modality mr images. IEEE Trans Image Process 29:9032–9043
Zhang D, Huang G, Zhang Q, Han J, Han J, Yu Y (2021) Cross-modality deep feature learning for brain tumor segmentation. Pattern Recogn 110(107):562
Zhang J, Cui Y, Lu S, Xiao L (2017) Multilayer image segmentation based on gaussian weighted euclidean distance and nonlinear interpolation. In: 2017 10Th international congress on image and signal processing, biomedical engineering and informatics (CISP-BMEI), pp 1–5. https://doi.org/10.1109/CISP-BMEI.2017.8302195
Zheng H, Chen Y, Yue X, Ma C, Liu X, Yang P, Lu J (2020) Deep pancreas segmentation with uncertain regions of shadowed sets. Magn Reson Imaging 68:45–52
Zhou Y, Li Y, Zhang Z, Wang Y, Wang A, Fishman EK, Yuille AL, Park S (2019) Hyper-pairing network for multi-phase pancreatic ductal adenocarcinoma segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 155–163
Zhou Y, Xie L, Fishman EK, Yuille AL (2017) Deep supervision for pancreatic cyst segmentation in abdominal ct scans. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 222–230
Zhu Z, Liu C, Yang D, Yuille A, Xu D (2019) V-nas: Neural architecture search for volumetric medical image segmentation. In: 2019 International conference on 3d vision (3DV), pp 240–248. https://doi.org/10.1109/3DV.2019.00035
Zhu Z, Xia Y, Xie L, Fishman EK, Yuille AL (2019) Multi-scale coarse-to-fine segmentation for screening pancreatic ductal adenocarcinoma. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 3–12
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 92046008, 62173252), the Shanghai Innovation Action Project of Science and Technology (No. 20Y11912500), and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, X., Chen, Y., Yue, X. et al. Local linear embedding based interpolation neural network in pancreatic tumor segmentation. Appl Intell 52, 8746–8756 (2022). https://doi.org/10.1007/s10489-021-02847-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02847-9