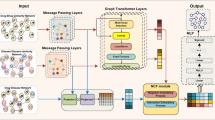
Abstract
Treating patients with complex diseases or co-existing conditions by polypharmacy (i.e., the use of drug combination) is very common. However, due to drug-drug interactions, polypharmacy often results in unpredictable side effects, which may endanger patients’ life. Moreover, since adverse drug reactions are rare, discovering polypharmacy side effects only from sparse drug-drug interactions remains challenging. Thus, it is necessary to explore the knowledge of polypharmacy side effects from the interaction network with complex relationships. In this paper, we propose a novel multi-modal graph convolutional neural network (M2GCN) for link prediction in multi-modal networks which consist of protein-protein interactions, drug-protein interactions and drug-drug interactions. Specifically, we first propose a propagation strategy to perform graph aggregations on each subgraph. Then we leverage consistency regularization to align the consistency across different subgraphs. Finally, referring to the DistMult method, we use the embeddings obtained above to calculate the probability of side effects between drugs. Experimental results on benchmark dataset show that our method significantly outperforms the compared network embedding models.







Similar content being viewed by others
References
Xu H, Sang S, Lu H (2020) Tri-graph information propagation for polypharmacy side effect prediction. arXiv:2001.10516
Tatonetti N P, Patrick P Y, Daneshjou R, Altman R B (2012) Data-driven prediction of drug effects and interactions. Science Translational Medicine, 125ra31–125ra31
Bansal M, Yang J, Karan C, Menden M P, Costello J C, Tang H, Xiao G, Li Y, Allen J, Zhong R et al (2014) A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol, 1213–1222
Bowes J, Brown A J, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S et al (2012) Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nature Reviews Drug Discovery, 909–922
Cheng Y, Gong Y, Liu Y, Song B, Zou Q (2021) Molecular design in drug discovery: a comprehensive review of deep generative models. Brief Bioinform
Ibrahim M M (2006) Ras inhibition in hypertension. J Hum Hypertens 20(2):101–108
Sun M, Zhao S, Gilvary C, Elemento O, Zhou J, Wang F (2020) Graph convolutional networks for computational drug development and discovery. Briefings in Bioinformatics
Song B, Li F, Liu Y, Zeng X (2021) Deep learning methods for biomedical named entity recognition: a survey and qualitative comparison. Brief Bioinform
Wishart D S, Feunang Y D, Guo A C, Lo E J, Marcu A, Grant J R et al (2018) Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Research, D1074–D1082
Szklarczyk D, Santos A, Von Mering C, Jensen L J, Bork P, Kuhn M (2016) Stitch 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Research, D380–D384
Zitnik M, Agrawal M, Leskovec J (2018) Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics, i457–i466
Kipf T N, Welling M (2017) Semi-supervised classification with graph convolutional networks. In: International conference on learning representations (ICLR)
Veličković P, Cucurull G, Casanova A, Romero A, Liò P, Bengio Y (2018) Graph attention networks. In: International conference on learning representations
Feng W, Zhang J, Dong Y, Han Y, Luan H, Xu Q et al (2020) Graph random neural network for semi-supervised learning on graphs. In: NeurIPS’20
Zhu Z, Fan X, Chu X, Bi J (2020) Hgcn: a heterogeneous graph convolutional network-based deep learning model toward collective classification. In: Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining, pp 1161–1171
Fu X, Zhang J, Meng Z, King I (2020) Magnn: metapath aggregated graph neural network for heterogeneous graph embedding. In: Proceedings of the web conference 2020, pp 2331–2341
Wang X, Ji H, Shi C, Wang B, Ye Y et al (2019) Heterogeneous graph attention network. In: The World wide web conference, pp 2022–2032
Dong Y, Chawla N V, Swami A (2017) metapath2vec: scalable representation learning for heterogeneous networks. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, pp 135–144
Shi C, Hu B, Zhao W X, Philip S Y (2018) Heterogeneous information network embedding for recommendation. IEEE Trans Knowl Data Eng
Wang X, Zhang Y, Shi C (2019) Hyperbolic heterogeneous information network embedding. In: Proceedings of the AAAI conference on artificial intelligence, pp 5337–5344
Yang Y, Guan Z, Li J, Zhao W, Cui J, Wang Q (2021) Interpretable and efficient heterogeneous graph convolutional network. IEEE Trans Knowl Data Eng
Zhao L, Akoglu L (2019) Pairnorm: tackling oversmoothing in gnns. In: International conference on learning representations
Li G, Muller M, Thabet A, Ghanem B (2019) Deepgcns: can gcns go as deep as cnns?. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 9267–9276
Ryall K A, Tan A C (2015) Systems biology approaches for advancing the discovery of effective drug combinations. Journal of Cheminformatics, 1–15
Loewe S (1953) The problem of synergism and antagonism of combined drugs. Arzneimittelforschung, 285–290
Lewis R, Guha R, Korcsmaros T, Bender A (2015) Synergy maps: exploring compound combinations using network-based visualization. Journal of Cheminformatics, 1–11
Takeda T, Hao M, Cheng T, Bryant S H, Wang Y (2017) Predicting drug–drug interactions through drug structural similarities and interaction networks incorporating pharmacokinetics and pharmacodynamics knowledge. Journal of Cheminformatics, 1–9
Sun Y, Sheng Z, Ma C, Tang K, Zhu R, Wu Z et al (2015) Combining genomic and network characteristics for extended capability in predicting synergistic drugs for cancer. Nature Communications, 1–10
Zitnik M, Zupan B (2016) Collective pairwise classification for multi-way analysis of disease and drug data. In: Biocomputing 2016: Proceedings Of The Pacific Symposium. World Scientific, pp 81–92
Chen X, Ren B, Chen M, Wang Q, Zhang L, Yan G (2016) Nllss: predicting synergistic drug combinations based on semi-supervised learning. PLoS Computational Biology, e1004975
Shi J-Y, Li J-X, Gao K, Lei P, Yiu S-M (2017) Predicting combinative drug pairs towards realistic screening via integrating heterogeneous features. BMC Bioinformatics, 1–9
Lin X, Quan Z, Wang Z-J, Ma T, Zeng X (2020) Kgnn: knowledge graph neural network for drug-drug interaction prediction. In: IJCAI, pp 2739–2745
Yu Z, Huang F, Zhao X, Xiao W, Zhang W (2021) Predicting drug–disease associations through layer attention graph convolutional network. Brief Bioinform, bbaa243
Lee C Y, Chen Y-P P (2021) Prediction of drug adverse events using deep learning in pharmaceutical discovery. Briefings in Bioinformatics, 1884–1901
Kastrin A, Ferk P, Leskošek B (2018) Predicting potential drug-drug interactions on topological and semantic similarity features using statistical learning. PloS One, e0196865
Bang S, Ho Jhee J, Shin H (2021) Polypharmacy side effect prediction with enhanced interpretability based on graph feature attention network. Bioinformatics
Huang K, Xiao C, Hoang T, Glass L, Sun J (2020) Caster: predicting drug interactions with chemical substructure representation. In: Proceedings of the AAAI conference on artificial intelligence, pp 702–709
Fu T, Xiao C, Qian C, Glass L M, Sun J (2021) Probabilistic and dynamic molecule-disease interaction modeling for drug discovery. In: Proceedings of the 27th ACM SIGKDD conference on knowledge discovery & data mining, pp 404–414
Defferrard M, Bresson X, Vandergheynst P (2016) Convolutional neural networks on graphs with fast localized spectral filtering. Advances in Neural Information Processing Systems, 3844–3852
Hamilton W L, Ying R, Leskovec J (2017) Inductive representation learning on large graphs. In: Proceedings of the 31st international conference on neural information processing systems, pp 1025–1035
Hu Z, Dong Y, Wang K, Sun Y (2020) Heterogeneous graph transformer. In: Proceedings of the web conference 2020, pp 2704–2710
Chen J, Huang F, Peng J (2021) Msgcn: multi-subgraph based heterogeneous graph convolution network embedding. Appl Sci
Chen J, Zhang A (2020) Hgmf: heterogeneous graph-based fusion for multimodal data with incompleteness. In: Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining, pp 1295–1305
Wang P, Agarwal K, Ham C, Choudhury S, Reddy C K (2021) Self-supervised learning of contextual embeddings for link prediction in heterogeneous networks. In: Proceedings of the web conference 2021, pp 2946–2957
Chen Y, Ma T, Yang X, Wang J, Song B, Zeng X (2021) Muffin: multi-scale feature fusion for drug–drug interaction prediction. Bioinformatics
Dai Y, Guo C, Guo W, Eickhoff C (2021) Drug–drug interaction prediction with wasserstein adversarial autoencoder-based knowledge graph embeddings. Briefings in Bioinformatics, bbaa256
Zeng X, Tu X, Liu Y, Fu X, Su Y (2022) Toward better drug discovery with knowledge graph. Current opinion in structural biology
Mikolov T, Sutskever I, Chen K, Corrado G S, Dean J (2013) Distributed representations of words and phrases and their compositionality. In: Advances in neural information processing systems, pp 3111–3119
Liu Q, Long C, Zhang J, Xu M, Lv P (2021) Triatne: tripartite adversarial training for network embeddings. IEEE Transactions on Cybernetics
Li Q, Wu X-M, Liu H, Zhang X, Guan Z (2019) Label efficient semi-supervised learning via graph filtering. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 9582–9591
Miao X, Gürel N M, Zhang W, Han Z, Li B, Min W et al (2019) Degnn: characterizing and improving graph neural networks with graph decomposition. arXiv:1910.04499
Wu F, Souza A, Zhang T, Fifty C, Yu T, Weinberger K (2019) Simplifying graph convolutional networks. In: International conference on machine learning. PMLR, pp 6861–6871
Yang B, Yih W-, He X, Gao J, Deng L (2014) Embedding entities and relations for learning and inference in knowledge bases. arXiv:1412.6575
Kuhn M, Letunic I, Jensen L J, Bork P (2016) The sider database of drugs and side effects. Nucleic Acids Research
Menche J, Sharma A, Kitsak M, Ghiassian S D, Vidal M, Loscalzo J, Barabási A-L (2015) Uncovering disease-disease relationships through the incomplete interactome. Science
Chatr-Aryamontri A, Breitkreutz B-J, Oughtred R, Boucher L, Heinicke S, Chen D et al (2015) The biogrid interaction database: 2015 update. Nucleic acids research
Szklarczyk D, Morris J H, Cook H, Kuhn M, Wyder S, Simonovic M et al (2016) The string database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research
Rolland T, Taşan M, Charloteaux B, Pevzner S J, Zhong Q, Sahni N et al (2014) A proteome-scale map of the human interactome network. Cell
Perozzi B, Al-Rfou R, Skiena S (2014) Deepwalk: online learning of social representations. In: Proceedings of the 20th ACM SIGKDD international conference on knowledge discovery and data mining, pp 701–710
Grover A, Leskovec J (2016) node2vec: scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, pp 855–864
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China under Grant (61906174, 62036010, 61903341, 61972362), in part by the China Postdoctoral Science Foundation under Grant 2020M672275, in part by the Department of Science and Technology of Henan Province under Grant (222102210248, 201100312000), in part by the Henan Province Natural Science Foundation under Grant (212300410291, 202300410378).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Q., Yao, E., Liu, C. et al. M2GCN: multi-modal graph convolutional network for modeling polypharmacy side effects. Appl Intell 53, 6814–6825 (2023). https://doi.org/10.1007/s10489-022-03839-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-03839-z