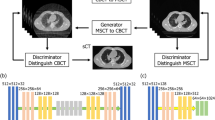
Abstract
Computed tomography (CT) plays key roles in radiotherapy treatment planning and PET attenuation correction (AC). Magnetic resonance (MR) imaging has better soft tissue contrast than CT and has no ionizing radiation but cannot directly provide information about photon interactions with tissue that is needed for radiation treatment planning and AC. Therefore, estimating synthetic CT (sCT) images from corresponding MR images and obviating CT scanning is of great interest, but can be particularly challenging in the abdomen owing to a range of tissue types and physiologic motion. For this purpose, inspired by deep learning, we design a novel generative adversarial network (GAN) model that organically combines ResNet, U-net, and auxiliary classifier-augmented GAN (RU-ACGAN for short). The significance of our effort is three-fold: 1) The combination of ResNet and U-net, instead of only the U-net which was commonly used in existing conditional GAN, is enlisted to constitute the generative network in RU-ACGAN. This has the potential to generate more accurate CT than existing methods. 2) Adding the classifier to the discriminant network makes the training process of the proposed model more stable, and thereby benefits the robustness of sCT estimation. 3) Owing to the delicate architecture, RU-ACGAN is capable of estimating superior sCT using only a limited quantity of training data. The experimental studies on ten subjects’ MR-CT pair images indicate that the proposed RU-ACGAN model can capture the potential, non-linear matching between the MR and CT images, and thus achieves the better performance for sCT estimation for the abdomen than many other existing methods.
Similar content being viewed by others
References
Brenner, D.J., Hall, E.J.: Computed tomography—an increasing source of radiation exposure. N. Engl. J. Med. 357(22), 2277–2284 (2007)
Stoessl, A.J.: Developments in neuroimaging: positron emission tomography. Parkinsonism Relat. Disord. 20(20S1), S180–S183 (2014)
Beyer, T., Townsend, D.W., Brun, T., Kinahan, P.E., Charron, M., Roddy, R., Jerin, J., Young, J., Byars, L., Nutt, R.: A combined PET/CT scanner for clinical oncology. J. Nucl. Med. 41(8), 1369–1379 (2000)
Shankar, L.K., Hoffman, J.M., Bacharach, S., Graham, M.M., Karp, J., Lammertsma, A.A., Larson, S., Mankoff, D.A., Siegel, B.A., Van den Abbeele, A., Yap, J., Sullivan, D.: Consensus recommendations for the use of 18F-FDG PET as an indicator of therapeutic response in patients in National Cancer Institute trials. J. Nucl. Med. 47(6), 1059–1066 (2016)
FDG-PET/CT Technical Committee: QIBA Profile: FDG-PET/CT as an Imaging Biomarker Measuring Response to Cancer Therapy Profile (V 1.05) (2013)
Dickson, J.C., O'Meara, C., Barnes, A.: A comparison of CT- and MR-based attenuation correction in neurological PET. Eur. J. Nucl. Med. Mol. Imaging. 41(6), 1176–1189 (2014)
Keereman, V., Fierens, Y., Broux, T., De Deene, Y., Lonneux, M., Vandenberghe, S.: MRI-based attenuation correction for PET/MRI using ultrashort echo time sequences. J. Nucl. Med. 51(5), 812–818 (2010)
Navalpakkam, B.K., Braun, H., Kuwert, T., Quick, H.H.: Magnetic resonance-based attenuation correction for PET/MR hybrid imaging using continuous valued attenuation maps. Investig. Radiol. 48(5), 323–332 (2013)
Hitz, S., Habekost, C., Furst, S., Delso, G., Forster, S., Ziegler, S., Nekolla, S.G., Souvatzoglou, M., Beer, A.J., Grimmer, T., Eiber, M., Schwaiger, M., Drzezga, A.: Systematic comparison of the performance of integrated whole-body PET/MR imaging to conventional PET/CT for 18F-FDG brain imaging in subjects examined for suspected dementia. J. Nucl. Med. 55(6), 923–931 (2014)
Berker, Y., Franke, J., Salomon, A., Palmowski, M., Donker, H.C., Temur, Y., Mottaghy, F.M., Kuhl, C., Izquierdo-Garcia, D., Fayad, Z.A., Kiessling, F., Schulz, V.: MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echo-time/Dixon MRI sequence. J. Nucl. Med. 53(5), 796–804 (2012)
Schramm, G., Langner, J., Hofheinz, F., Petr, J., Beuthien-Baumann, B., Platzek, I., Steinbach, J., Kotzerke, J., van den Hoff, J.: Erratum to: Quantitative accuracy of attenuation correction in the Philips Ingenuity TF whole-body PET/MR system: a direct comparison with transmission-based attenuation correction. Magnetic Resonance Mater. Phys. Biol. Med. 28(1), 101 (2015)
Schramm, G., Langner, J., Hofheinz, F., Petr, J., Beuthien-Baumann, B., Platzek, I., Steinbach, J., Kotzerke, J., van den Hoff, J.: Quantitative accuracy of attenuation correction in the Philips Ingenuity TF whole-body PET/MR system: a direct comparison with transmission based attenuation correction. Magnetic Resonance Materials in Physics, Biology and Medicine. 26(1), 115–126 (2013)
Samarin, A., Burger, C., Wollenweber, S.D., Crook, D.W., Burger, I.A., Schmid, D.T., von Schulthess, G.K., Kuhn, F.P.: PET/MR imaging of bone lesions--implications for PET quantification from imperfect attenuation correction. Eur. J. Nucl. Med. Mol. Imaging. 39(7), 1154–1160 (2012)
Arabi, H., Rager, O., Alem, A., Varoquaux, A., Becker, M., Zaidi, H.: Clinical assessment of MR-guided 3-class and 4-class attenuation correction in PET/MR. Mol. Imaging Biol. 17(2), 264–276 (2015)
Aznar, M.C., Sersar, R., Saabye, J., Ladefoged, C.N., Andersen, F.L., Rasmussen, J.H., Löfgren, J., Beyer, T.: Whole-body PET/MRI: the effect of bone attenuation during MR-based attenuation correction in oncology imaging. Eur. J. Radiol. 83(7), 1177–1183 (2014)
Izquierdo-Garcia, D., Sawiak, S.J., Knesaurek, K., Narula, J., Fuster, V., Machac, J., Fayad, Z.A.: Comparison of MR-based attenuation correction and CT-based attenuation correction of whole-body PET/MR imaging. Eur. J. Nucl. Med. Mol. Imaging. 41(8), 1574–1584 (2014)
Bezrukov, I., Schmidt, H., Mantlik, F., Schwenzer, N., Brendle, C., Scholkopf, B., Pichler, B.J.: MR-based attenuation correction methods for improved PET quantification in lesions within bone and susceptibility artifact regions. J. Nucl. Med. 54(10), 1768–1774 (2013)
Hofmann, M., Pichler, B., Schölkopf, B., et al.: Towards quantitative PET/MRI: a review of MR-based attenuation correction techniques. European Journal of Nuclear Medicine & Molecular Imaging. 36(1), 93–104 (2009)
Sjölund, J., Forsberg, D., Andersson, M., Knutsson, H.: Generating subject specific pseudo-CT of the head from MR using atlas-based regression. Phys. Med. Biol. 60(2), 825–839 (2015)
Dowling, J.A., Lambert, J., Parker, J., Salvado, O., Fripp, J., Capp, A., Wratten, C., Denham, J.W., Greer, P.B.: An atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 83(1), e5–e11 (2012)
Su, K.H., Hu, L., Stehning, C., Helle, M., Qian, P., Thompson, C.L., Pereira, G.C., Jordan, D.W., Herrmann, K.A., Traughber, M., Muzic Jr., R.F., Traughber, B.J.: Generation of brain pseudo-CTs using an undersampled, single-acquisition UTE-mDixon pulse sequence and unsupervised clustering. Medical Physics. 42(8), 4974–4986 (2015)
Hsu, S., Cao, Y., Balter, J.: MO-G-BRA-02: Investigation of a method for generating synthetic CT models from MRI scans for radiation therapy. Med. Phys. 39(6Part22), 3881–3881 (2012)
Khalifé, M., Fernandez, B., Jaubert, O., Soussan, M., Brulon, V., Buvat, I., Comtat, C.: Subject-specific bone attenuation correction for brain PET/MR: can ZTE-MRI substitute CT scan accurately? Phys. Med. Biol. 62(19), 7814–7832 (2017)
Jog, A., Carass, A., Prince, J.L.: Improving magnetic resonance resolution with supervised learning. In: Proc IEEE 11th International Symposium on Biomedical Imaging, pp. 987–990 (2014)
Huynh, T., Gao, Y., Kang, J., Wang, L., Zhang, P., Lian, J., Shen, D.: Estimating CT image from MRI data using structured random forest and auto-context model. IEEE Trans. Med. Imaging. 35(1), 174–183 (2015)
Dowling, J.A., Sun, J., Pichler, P., Rivest-Hénault, D., Ghose, S., Richardson, H., FRANZCR, C.W., Martin, J., Arm, J., Best, L., Chandra, S.S., Fripp, J., Menk, F.W., Greer, P.B.: Automatic substitute computed tomography generation and contouring for magnetic resonance imaging (MRI)-alone external beam radiation therapy from standard MRI sequences. Int. J. Radiat. Oncol. Biol. Phys. 93(5), 1144–1153 (2015)
Qian, P., Chen, Y., Kuo, J.W., Zhang, Y.D., Jiang, Y., Zhao, K., Helo, R.A., Friel, H., Baydoun, A., Zhou, F., Heo, J.U., Avril, N., Herrmann, K., Ellis, R., Traughber, B., Jones, R.S., Wang, S., Su, K.H., Muzic Jr., R.F.: mDixon-based synthetic CT generation for PET attenuation correction on abdomen and pelvis jointly using transfer fuzzy clustering and active learning-based classification. IEEE Trans. Med. Imaging. (2019). https://doi.org/10.1109/TMI.2019.2935916
Johnstone, E., Wyatt, J.J., Henry, A.M., Short, S.C., Sebag-Montefiore, D., Murray, L., Kelly, C.G., McCallum, H.M., Speight, R.: Systematic review of synthetic computed tomography generation methodologies for use in magnetic resonance imaging-only radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 100(1), 199–217 (2018)
Lecun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature. 521, 436–444 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Odena, A., Olah, C., Shlens, J.: Conditional image synthesis with auxiliary classifier GANs. In: Proceedings of the 34th International Conference on Machine Learning, 70, pp. 2642–2651 (2017)
Reed, S., Akata, Z., Yan, X., Logeswaran, L.: Generative adversarial text to image synthesis. In: Proceedings of the 33rd International Conference on Machine Learning, 48, pp. 1060–1069 (2016)
Narasimha, R., Fern, X.Z., Raich, R.: Simultaneous segmentation and classification of bird song using CNN. In: 2017 IEEE international conference on acoustics, Speech Signal Process, vol. 2017. https://doi.org/10.1109/ICASSP.2017.7952135
Nauman, M., Rehman, H.U., Politano, G., Benso, A.: Beyond homology transfer: deep learning for automated annotation of proteins. J. Grid Comput. 17, 225–237 (2019)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(4), 640–651 (2014)
Chen, H., Qi, X., Yu, L., Heng, P.A.: DCAN: Deep contour-aware networks for accurate gland segmentation. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition, pp. 2487–2496 (2016)
Kitchen, A., Seah, J.: Deep generative adversarial neural networks for realistic prostate lesion MRI synthesis. arXiv. 1708.00129 (2017)
Kohl, S., Bonekamp, D., Schlemmer, H.P., Yaqubi, K., Hohenfellner, M., Hadaschik, B., Radtke, J.P., Maier-Hein, K.: Adversarial networks for the detection of aggressive prostate cancer. arXiv. 1702.08014 (2017)
Fu, J., Yang, Y., Singhrao, K., Ruan, D., Chu, F.I., Low, D.A., Lewis, J.H.: Deep learning approaches using 2D and 3D convolutional neural networks for generating male pelvic synthetic computed tomography from magnetic resonance imaging. Med. Phys. (2019). https://doi.org/10.1002/mp.13672
Emami, H., Dong, M., Nejad-Davarani, S.P., Glide-Hurst, C.K.: Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med. Phys. 45(8), 3627–3636 (2018)
Han, X.: MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 44(4), 1408–1419 (2017)
Zia, T., Razzaq, S.: Residual recurrent highway networks for learning deep sequence prediction models. J. Grid Comput. 1–8 (2018). https://doi.org/10.1007/s10723-018-9444-4
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: MICCAI 2015, pp. 234–241 (2015)
Mirza, M., Osindero, S.: Conditional generative adversarial nets. arXiv. 1411.1784 (2014)
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville, A., Bengio, Y.: Generative adversarial nets. In: Proceedings of the 27th International Conference on Neural Information Processing Systems. 2, pp. 2672–2680 (2014)
Radford, A., Metz, L., Chintala, S.: Unsupervised representation learning with deep convolutional generative adversarial networks. In: 2015 ICLR (2015)
Arjovsky, M., Chintala, S., Bottou, L.: Wasserstein GAN. arXiv. 1701.07875 (2017)
Isola, P., Zhu, J.Y., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition, pp. 5967–5976 (2017)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd International Conference on Machine Learning, 37, pp. 448–456 (2015)
Xu, B., Wang, N., Chen, T., Li, M.: Empirical evaluation of rectified activations in convolutional network. arXiv. 1505.00853 (2015)
Gates Jr., G.H., Merkle, L.D., Lamont, G., Pachter, R.: Simple genetic algorithm parameter selection for protein structure prediction. In: Proceedings of 1995 IEEE International Conference on Evolutionary Computation (2002). https://doi.org/10.1109/ICEC.1995.487455
Gkoutioudi, K.Z., Karatza, H.D.: Multi-criteria job scheduling in grid using an accelerated genetic algorithm. J. Grid Comput. 10, 311–323 (2012)
Khajemohammadi, H., Fanian, A., Gulliver, T.A.: Efficient workflow scheduling for grid computing using a leveled multi-objective genetic algorithm. J. Grid Comput. 12, 637–663 (2014)
Su, K.H., Friel, H.T., Kuo, J.W., Helo, R.A., Baydoun, A., Stehning, C., Crisan, A.N., Devaraj, A., Jordan, D.W., Qian, P., Leisser, A., Ellis, R.J., Herrmann, K.A., Avril, N., Traughber, B.J., Muzic Jr., R.F.: UTE-mDixon-based thorax synthetic CT generation. Med. Phys. 46(8), 3520–3531 (2019)
Janssens, G., Jacques, L., de Xivry, J.O., Geets, X., Macq, B.: Diffeomorphic registration of images with variable contrast enhancement. Int. J. Biomed. Imaging. 2011, Art. ID 891585 (2011)
Qian, P., Sun, S., Jiang, Y., Su, K.-H., Ni, T., Wang, S., Muzic Jr., R.F.: Cross-domain, soft-partition clustering with diversity measure and knowledge reference. Pattern Recogn. 50, 155–177 (2016)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: Proceedings of the 3rd International Conference on Learning Representations (2015)
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under Grants 61772241 and 61702225, by the Natural Science Foundation of Jiangsu Province under Grant BK20160187, by the Fundamental Research Funds for the Central Universities under Grant JUSRP51614A, by 2016 Qinglan Project of Jiangsu Province, by 2016 Six Talent Peaks Project of Jiangsu Province, and by the Science and Technology Demonstration Project of Social Development of Wuxi under Grant WX18IVJN002. Research in this publication was also supported by National Cancer Institute of the National Institutes of Health, USA, under award number R01CA196687 (The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, USA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qian, P., Xu, K., Wang, T. et al. Estimating CT from MR Abdominal Images Using Novel Generative Adversarial Networks. J Grid Computing 18, 211–226 (2020). https://doi.org/10.1007/s10723-020-09513-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10723-020-09513-3