Abstract
We present a new approach to clustering and visualization of the DNA microarray gene expression data. We utilize the self-organizing map (SOM) framework for handling (dis)similarities between genes in terms of their expression characteristics. We rely on appropriately defined distances between ranked genes-attributes, also capable of handling missing values. As a case study, we consider breast cancer data and the gene ESR1, whose expression alterations, appearing for many of the tumor subtypes, have been already observed to be correlated with some other significant genes. Preliminary results positively verify applicability of our approach, although further development is definitely needed. They suggest that it may be very effective when used by the domain experts. The algorithmic toolkit is enriched with GUI enabling the users to interactively support the SOM optimization process. Its effectiveness is achieved by drag&drop techniques allowing for the cluster modification according to the expert knowledge or intuition.
Similar content being viewed by others
References
Aas T, Botstein D, Brown P. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS 2001;98:10869–10874.
Akslen L, Botstein D, Eisen M, Fluge O, Jeffrey S, Lonning P. Molecular portraits of human breast tumors. Nature 2000;406:747–752.
Altman R, Botstein D, Brown P, Cantor M, Hastie T, Tibshirani R. Missing value estimation methods for dna microarrays. Bioinformatics 2001;17:520–525.
Anders K, Botstein D, Brown P. Comprehensive identification of cell cycle-regulated genes of the yeast saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 1998;9:3273–3297.
Boyapati S, Shu X, Ruan Z. Polymorphisms in er-alpha gene interact with estrogen receptor status in breast cancer survival. Clin Cancer Res 2005;11:1093–1098.
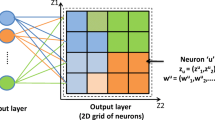
Castrn E, Kaski S, Nikkil J, Trrnen P, Wong G. Analysis and visualization of gene expression data using self-organizing maps. In: IEEE - EURASIP Workshop on Nonlinear Signal and Image Processing (NSIP-01), Baltimore 2001.
de Brevern A, Hazout S, Malpertuy A. Influence of microarrays experiments missing values on the stability of gene groups by hierarchical clustering. BMC Bioinformatics 2004;5:114.
Demeter J, Deng S, Geisler S. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003;100:8418–8423.
Eriksen K, Hornquist M, Sneppen K. Visualization of large-scale correlations in gene expressions. Funct Integr Genomics 2004;4:241–245.
Friedman N, Linial M, Nachman I, Pe'er D. Using bayesian networks to analyze expression data. Journal of Computational Biology 2000;7:601–620.
Gruźdź A, Ihnatowicz A, Ślezak D. Gene expression clustering: Dealing with the missing values. In: Klopotek, M.A., Trojanowski, K., and Wierzchoń, S., eds., Proc. of IIS 2005, LNAI, Springer Verlag, 2005a; 521–530.
Gruźdź A, Ihnatowicz A, Ślezak D. Interactive som-based gene grouping: An approach to gene expression data analysis. In Hacid, M.-S., Murray, N.V., Raś, Z.W., and Tsumoto, S., eds. Proc. of ISMIS 2005, LNAI, Springer Verlag 2005b; 514–523.
Ishii S, Matsubara K, Monden M. A bayesian missing value estimation method. Bioinformatics 2003;19:2088–2096.
Kapur J, Kesavan H. Entropy Optimization Principles with Applications. Academic Press, 1992.
Kaski S. Som-based exploratory analysis of gene expression data. In: Advances in Self-Organizing Maps, Springer Verlag 2001; 124–131.
Kohonen T. Self-organized formation of topologically correct feature maps. Biological Cybernetics 1982;43:59–69.
Lacroix M, Leclercq G. About gata3, hnf3a and xbp1, three genes co-expressed with the oestrogen receptor-gene (esr1) in breast cancer. Molecular and Cellular Endocrinology 2004;219:1–7.
Lawrence C, Liu J, Palumbo M, Zhang J. Bayesian clustering with variable and transformation selections. In: Bayesian Statistics 7, Oxford University Press. 2003; 249–275.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gruźdź, A., Ihnatowicz, A. & Ślʁzak, D. Interactive Gene Clustering—A Case Study of Breast Cancer Microarray Data. Inf Syst Front 8, 21–27 (2006). https://doi.org/10.1007/s10796-005-6100-x
Issue Date:
DOI: https://doi.org/10.1007/s10796-005-6100-x