Abstract
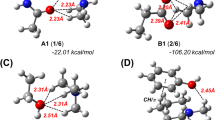
Using Density Functional Theory, the hydrogen bonding energy is calculated for the interaction of phenol and aniline with four model compounds representing the protein backbone and various amino acid site chain residues. The models are methanol, protonated methylamine, formaldehyde and acetate anion. The H-bond energies for the uncharged species are ∼2.5kcalmol−1, whereas the charged model compounds bind with much higher energies of ∼20kcalmol−1. The effect of para-substitution on the hydrogen bond energies is determined. Substitution has little effect on the H-bond energy of the neutral complexes (<2kcalmol−1), but for the positively and negatively charged systems substitution drastically alters the binding energies, e.g., 14.3kcalmol−1 for para-NO2. In the context of protein–ligand binding, relatively small changes in binding energy can cause large changes in affinity due to their exponential relationship. This means that for –NO2 an enormous change of 10 orders of magnitude for the affinity constant is predicted. These calculations allow prediction of H-bonds, using different substituents, in order to fine-tune and optimize ligand–protein interactions in the search for drug candidates.
Similar content being viewed by others
References
Pauling, L., Corey, R.B. and Branson, H.R., Proc. Natl. Acad. Sci. USA, 37 (1951) 205.
Pauling, L. and Corey, R.B., Proc. Natl. Acad. Sci. USA, 37 (1951) 729.
Böhm, H.J. and Klebe, G., Angew. Chem. Int. Ed. Engl., 35 (1996) 2588.
Klebe, G. and Böhm, H.J., Recept. Signal Transduction Res., 17 (1997) 459.
Höltje, H.D., Sippl, W., Rognan, D. and Folkers, G., Molecular Modeling, Second Edition. Wiley-VCH, Weinheim, Germany, 2003.
An analog of the highly successful Glivec kinase inhibitor interacts with the protein through hydrogen bonds, some of which confer specificity [7].
Schindler, T., Bornmann, W., Pellicena, P., Miller, W.T., Clarkson, B. and Kuriyan, J., Science, 289 (2000) 1938.
Very weak correlation has been found between the number of contributing H-bonds to binding affinity [3].
Ward, W.H.J. and Holdgate, G.A., In King, F.D. and Oxford, A.W. (Eds.), Progress in Medicinal Chemistry, Vol. 38.Elsevier Science, 2001, pp. 309-376.
Roberts, D.A. and Ward, W.H.J., In King, F.D. (Ed.), Medicinal Chemistry:Principles and Practice, Royal Society of Chemistry, Cambridge, UK, 2002, pp. 64-90.
Abraham, M.H. and Platts, J.A., J. Org. Chem., 66 (2001) 3484.
Ahn, D., Park, S., Lee, S. and Kim, B., J. Phys. Chem. A, 107 (2003) 131.
Pejov, L., Chem.Phys., 285 (2002) 177.
Koch, W. and Holthausen, M.C., A Chemist 's Guide to Density Functional Theory. Wiley-VCH, Weinheim, Germany, 1999.
Reynisson, J. and Steenken, S., J. Mol. Struct. (Theochem), 635 (2003) 133.
Reynisson, J. and Steenken, S., Phys. Chem. Chem. Phys., 4 (2002) 5353.
Abdali, S., Jalkanen, K.J., Cao, X., Nafie, L.A. and Bohr, H., Phys. Chem. Chem. Phys., 6 (2004) 2434.
Tuma, C. and Sauer, J., Chem. Phys. Lett., 387 (2004) 388.
Shishkin, O.V., Elstner, M., Frauenheim, T. and Suhai, S., Int. J. Mol. Sci., 4 (2003) 537.
Guerra, C.F., Bickelhaupt, F.M., Snijders, J.G. and Baerends, E.J., J. Am. Chem. Soc., 122 (2000) 4117.
Braida, B., Hiberty, P.C. and Savin, A., J. Phys. Chem. A, 102 (1998) 7872.
Pollet, R., Savin, A., Leininger, T. and Stoll, H., J. Chem. Phys., 116 (2002) 1250.
Kamiya, M., Tsuneda, T. and Hirao, K., J. Chem. Phys., 117 (2002) 6010.
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., J.A. Montgomery, J., Vreven, T., Kudin, K.N., Burant, J.C., Millam, J.M., Iyengar, S.S., Tomasi, J., Barone, V., Mennucci, B., Cossi, M., Scalmani, G., N. Rega, G.A. Petersson, Nakatsuji, H., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Klene, M., Li, X., Knox, J.E., Hratchian, H.P., Cross, J.B., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Ayala, P.Y., Morokuma, K., Voth, G.A., Salvador, P., Dannenberg, J.J., Zakrzewski, V.G., Dapprich, S., Daniels, A.D., Strain, M.C., Farkas, O., Malick, D.K., Rabuck, A.D., Raghavachari, K., Foresman, J.B., Ortiz, J.V., Cui, Q., Baboul, A.G., Clifford, S., Cioslowski, J., Stefanov, B.B., Liu, G., Liashenko, A., Piskorz, P., Komaromi, I., Martin, R.L., Fox, D.J., Keith, T., Al-Laham, M.A., Peng, C.Y., Nanayakkara, A., Challacombe, M., Gill, P.M.W., Johnson, B., Chen, W., Wong, M.W., Gonzalez, C. and Pople, J.A. GAUSSIAN 03 (B.3). Gaussian, Inc., Pittsburgh, PA, 2003.
Lee, C., Yang, W. and Parr, R.G., Phys. Rev. B., 37 (1988) 785.
Becke, A.D., Phys. Rev. A., 38 (1988) 3098.
Becke, A.D., J. Chem. Phys., 98 (1993) 5648.
Frisch, M.J., Pople, J.A. and Binkley, J.S., J. Chem. Phys., 80 (1984) 3265.
Wong, M.W., Chem. Phys. Lett., 256 (1996) 391.
Boys, S.F. and Bernardi, F., Mol. Phys., 19 (1970) 553.
Duijneveldt, F.B.v., Rijdt, J.G.C.M.v.D.-v.d. and Lenthe, J.H.v., Chem. Rev., 94 (1994) 1873.
It also applies to some extent to arginine. In general, we do not try to cover all of the amino acids but focus on the most interesting ones in our opinion.
Here the consequences on the entropy term are ignored in order to keep the model simple. The qualitative effects on entropy are discussed later in the paper.
Desiraju, G.R. and Steiner, T., The Weak Hydrogen Bond. University Press, Oxford, UK, 1999, p. 12.
Additionally, the aniline-acetate anion system, substituted with-CH2OH, forms bidentate bonding, i.e., each respective hydrogen on aniline forms a bond with each oxygen atom on the acetate molecule with H-bond lengths of 1.8 and 2.6 Å, respectively. This is the only complex, which forms a bidentate bonding observed in this work.
Reynisson, J. and Steenken, S., Org. Biomol. Chem., 2 (2004) 578.
Steenken, S., Chem.Rev., 89 (1989) 503.
Reynisson, J. and Steenken, S., Phys. Chem. Chem. Phys., 4 (2002) 5346.
Isaacs, N., Physical Organic Chemistry. Longman Scientic and Technical, Essex, UK, 1987, pp. 146-192.
The R values for the slopes lie between 0.97509 and 0.87004.
Nitro (σpara = 0.78), trifluoromethyl (σpara = 0.53) and methylsulphonite (σpara = 0.73).
Boström, J., Norrby, P. and Liljefors, T., J. Comput.-aided Mol. Des., 12 (1998) 383.
Vieth, M., Hirst, J.D. and Brooks III, C.L., J. Comput.-aided Mol. Des., 12 (1998) 563.
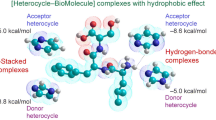
For intercalation of ligands into the DNA stack hydro-phobicity provides the overwhelming driving force for complex formation [45, 46].
Chaires, J.B., Curr. Opin. Struct. Biol., 8 (1998) 314.
Chaires, J.B., Biopolymers, 44 (1997) 201.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Reynisson, J., Mcdonald, E. Tuning of hydrogen bond strength using substituents on phenol and aniline: A possible ligand design strategy. J Comput Aided Mol Des 18, 421–431 (2004). https://doi.org/10.1007/s10822-004-3741-7
Issue Date:
DOI: https://doi.org/10.1007/s10822-004-3741-7