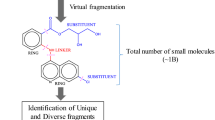
Abstract
We present a new algorithm for the enumeration of chemical fragment spaces under constraints. Fragment spaces consist of a set of molecular fragments and a set of rules that specifies how fragments can be combined. Although fragment spaces typically cover an infinite number of molecules, they can be enumerated in case that a physicochemical profile of the requested compounds is given. By using min–max ranges for a number of corresponding properties, our algorithm is able to enumerate all molecules which obey these properties. To speed up the calculation, the given ranges are used directly during the build-up process to guide the selection of fragments. Furthermore, a topology based fragment filter is used to skip most of the redundant fragment combinations. We applied the algorithm to 40 different target classes. For each of these, we generated tailored fragment spaces from sets of known inhibitors and additionally derived ranges for several physicochemical properties. We characterized the target-specific fragment spaces and were able to enumerate the complete chemical subspaces for most of the targets.







Similar content being viewed by others
Abbreviations
- ACE:
-
Angiotensin-converting enzyme
- AChE:
-
Acetylcholinesterase
- ADA:
-
Adenosine deaminase
- ALR2:
-
Aldose reductase
- AmpC:
-
AmpC β-lactamase
- AR:
-
Androgen receptor
- CDK2:
-
Cyclin-dependent kinase 2
- COMT:
-
Catechol O-methyltransferase
- COX-1:
-
Cyclooxygenase-1
- COX-2:
-
Cyclooxygenase-2
- DHFR:
-
Dihydrofolate reductase
- EGFr:
-
Epidermal growth factor receptor
- ER:
-
Estrogen receptor
- FGFr1:
-
Fibroblast growth factor receptor kinase
- FXa:
-
Factor Xa
- GART:
-
Glycinamide ribonucleotide transformylase
- GPB:
-
Glycogen phosphorylase β
- GR:
-
Glucocorticoid receptor
- HIVPR:
-
HIV protease
- HIVRT:
-
HIV reverse transcriptase
- HMGR:
-
Hydroxymethylglutaryl-CoA reductase
- HSP90:
-
Human heat shock protein 90
- InhA:
-
Enoyl ACP reductase
- MR:
-
Mineralocorticoid receptor
- NA:
-
Neuraminidase
- P38 MAP:
-
P38 mitogen activated protein
- PARP:
-
Poly(ADP-ribose) polymerase
- PDE5:
-
Phosphodiesterase 5
- PDGFrb:
-
Platelet derived growth factor receptor kinase
- PNP:
-
Purine nucleoside phosphorylase
- PPARg:
-
Peroxisome proliferator activated receptor γ
- PR:
-
Progesterone receptor
- RXRa:
-
Retinoic X receptor α
- SAHH:
-
S-adenosyl-homocysteine hydrolase
- SRC:
-
Tyrosine kinase SRC
- TK:
-
Thymidine kinase
- VEGFr2:
-
Vascular endothelial growth factor receptor
References
Walters WP, Stahl MT, Murcko MA (1994) Drug Discov Today 4:160
Oprea TI, Gottfries J (2001) J Comb Chem 3:157
Lipinski C, Hopkins A (2004) Nature 432:855
Dobson CM (2004) Nature 432:824
Rarey M, Degen J, Reulecke I (2007) Docking and scoring for structure-based drug design. Bioinformatics – from genomes to therapies, vol 2. Wiley-VCH Verlag GmbH & Co. KGaA, Weinmheim, p 541
Rarey M, Stahl M (2001) J Comput-Aided Mol Design 15:497
Schneider G, Lee M-L, Stahl M, Schneider P (2000) J Comput-Aided Mol Design 14:487
Lewell XQ, Judd DB, Watson SP, Hann MM (1998) J Chem Inf Comput Sci 38:511
WDI: World Drug Index, Derwent Information
Rarey M, Dixon JS (1998) J Comput-Aided Mol Design 12:471
Schneider G, Fechner U (2005) Nat Rev 4:649
Degen J, Rarey M (2006) ChemMedChem 1:854
Rarey M, Kramer B, Lengauer T, Klebe G (1996) J Mol Biol 261:470
Weber L (2005) QSAR Comb Sci 24:809
Ghose AK, Viswanadhan VN (2001) Combinatorial library design and evaluation. Marcel Dekker Inc
Knuth DE (1968) The Art of computer programming – fundamental algorithms. Addison-Wesley
Weininger D, Weininger A, (1986) J Chem Inf Comput Sci 29:97
Knott GD (1971) ACM 1:175
Bentley JL (1975) Commun ACM 18:509
Huang N, Shoichet BK, Irwin JJ (2006) J Med Chem 49:6789
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Adv Drug Deliv Rev 23:3
Wildman SA (1999) J Chem Inf Comput Sci 39:868
SYBYL mol2 file format. Tripos Inc
Sadowski J, Rudolph C, Gasteiger J (1990) Tetrahedron Comput Methodol 3:537
Sadowski J, Schwab CH, Gasteiger J (2006) Corina, version 3.10, Molecular Networks GmbH Computerchemie. Erlangen
Stahl M, Rarey M (2001) J Med Chem 44:1035
Fricker P, Gastreich M, Rarey M (2004) J Chem Inf Comput Sci 44:1065
Acknowledgments
The authors thank Andrea Zaliani (Lilly Research Laboratories, Hamburg, Germany) for careful reading of the manuscript and Tora Pommerencke, Christoph Müller and Florian Krull, who developed some useful ideas how to improve the performance of the enumeration during their time as project students. Additionally, we thank Patrick Maaß for help in generating the fragments out of the inhibitors sets. We further thank all partners in the NovoBench project for fruitful feedback and the german federal ministry of education and research for funding (grant 313324A).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pärn, J., Degen, J. & Rarey, M. Exploring fragment spaces under multiple physicochemical constraints. J Comput Aided Mol Des 21, 327–340 (2007). https://doi.org/10.1007/s10822-007-9121-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-007-9121-3