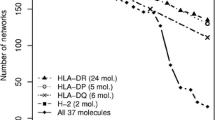
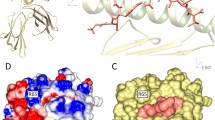
Abstract
T-cells recognize antigens via their T-cell receptors. The major histocompatibility complex (MHC) binds antigens in a specific way, transports them to the surface and presents the peptides to the TCR. Many in silico approaches have been developed to predict the binding characteristics of potential T-cell epitopes (peptides), with most of them being based solely on the amino acid sequence. We present a structural approach which provides insights into the spatial binding geometry. We combine different tools for side chain substitution (threading), energy minimization, as well as scoring methods for protein/peptide interfaces. The focus of this study is on high data throughput in combination with accurate results. These methods are not meant to predict the accurate binding free energy but to give a certain direction for the classification of peptides into peptides that are potential binders and peptides that definitely do not bind to a given MHC structure. In total we performed approximately 83,000 binding affinity prediction runs to evaluate interactions between peptides and MHCs, using different combinations of tools. Depending on the tools used, the prediction quality ranged from almost random to around 75% of accuracy for correctly predicting a peptide to be either a binder or a non-binder. The prediction quality strongly depends on all three evaluation steps, namely, the threading of the peptide, energy minimization and scoring.



Similar content being viewed by others
References
Saxova P, Buus S, Brunak S, Kesmir C (2003) Predicting proteasomal cleavage sites: a comparison of available methods. Int Immunol 15(7):781–787. doi:10.1093/intimm/dxg084
Korber B, LaBute M, Yusim K (2006) Immunoinformatics comes of age. PLOS Comput Biol 2(6):e71. doi:10.1371/journal.pcbi.0020071
Tsurui H, Takahashi T (2007) Prediction of T-cell epitope. J Pharmacol Sci 105(4):299–316. doi:10.1254/jphs.CR0070056
Sousa SF, Fernades P, Ramos MJ (2006) Protein-ligand docking current status and future challanges. Proteins 65:15–26. doi:10.1002/prot.21082
Gowthaman U, Agrewala JN (2008) In silico tools for predicting peptides binding to HLA-class II molecules: more confusion than conclusion. J Proteome Res 7(1):154–163. doi:10.1021/pr070527b
Miller PJ, Pazy Y, Conti B, Riddle D, Appella E, Collins EJ (2007) Single MHC mutation eliminates enthalpy associated with T cell receptor binding. J Mol Biol 373(2):315–327. doi:10.1016/j.jmb.2007.07.028
Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351:290–296. doi:10.1038/351290a0
Kjer-Nielsen L, Clements CS, Purcell AW et al (2003) A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity 18(1):53–64. doi:10.1016/S1074-7613(02)00513-7
Bergman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. doi:10.1093/nar/28.1.235
Peters B, Bui HH, Frankild S et al (2006) A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLOS Comput Biol 2(6):e65. doi:10.1371/journal.pcbi.0020065
Rognan D, Zimmermann N, Jung G, Folkers G (1992) Molecular dynamics study of a complex between the human histocompatibility antigen HLA-A2 and the IMP58–66 nonapeptide from influenza virus matrix protein. Eur J Biochem 208(1):101–113. doi:10.1111/j.1432-1033.1992.tb17163.x
Zoete V, Michielin O (2007) Comparison between computational alanine scanning and per-residue binding free energy decomposition for protein-protein association using MM-GBSA: Application to the TCR-p-MHC complex. Proteins 67(4):1026–1047. doi:10.1002/prot.21395
Gregoire C, Lin SY, Mazza G, Rebai N, Luescher IF, Malissen B (1996) Covalent assembly of a soluble T cell receptor-peptide-major histocompatibility class I complex. Proc Natl Acad Sci USA 93(14):7184–7189. doi:10.1073/pnas.93.14.7184
Toh H, Kamikawaji N, Tana T, Muta S, Sasazuki T, Kuhara S (2000) Magnitude of structural changes of the T-cell receptor binding regions determine the strength of T-cell antagonism: molecular dynamics simulations of HLA-DR4 (DRB1*0405) complexed with analogue peptide. Protein Eng 13(6):423–429. doi:10.1093/protein/13.6.423
Omasits U, Knapp B, Neumann M et al (2008) Analysis of key parameters for molecular dynamics of pMHC molecules. Mol Simul 34:781–793. doi:10.1080/08927020802256298
Wan S, Coveney P, Flower DR (2004) Large-scale molecular dynamics simulations of HLA-A*0201 complexed with a tumor-specific antigenic peptide: can the alpha3 and beta2 m domains be neglected? J Comput Chem 25(15):1803–1813. doi:10.1002/jcc.20100
Knapp B, Omasits U, Schreiner W (2008) Side chain substitution benchmark for peptide/MHC interaction. Protein Sci 17(6):977–982. doi:10.1110/ps.073402508
Canutescu AA, Shelenkov AA, Dunbrack RL Jr (2003) A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci 12(9):2001–2014. doi:10.1110/ps.03154503
Xu J (2005) Rapid side-chain prediction via tree decomposition. RECOMB 3500:423–439
Hartmann C, Antes I, Lengauer T (2007) IRECS: a new algorithm for the selection of most probable ensembles of side-chain conformations in protein models. Protein Sci 16(7):1294–1307. doi:10.1110/ps.062658307
Lindahl E, Hess B, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317
Rognan D, Lauemoller SL, Holm A, Buus S, Tschinke V (1999) Predicting binding affinities of protein ligands from three-dimensional models: application to peptide binding to class I major histocompatibility proteins. J Med Chem 42(22):4650–4658. doi:10.1021/jm9910775
Wang R, Lai L, Wang S (2002) Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des 16:11–26. doi:10.1023/A:1016357811882
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152. doi:10.1002/jcc.20634
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36
Spearman C (1904) The proof, measurement of association between two things. By C. Spearman, 1904. Am J Psychol 100(3–4):441–471
Roc-macro (2008) Nonparametric comparison of areas under correlated ROC curves. SAS website 2008 July 16. Available from http://support.sas.com/kb/25/017.html. Cited 2008 Jul 16
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3):837–845. doi:10.2307/2531595
Kates L, Petzoldt T proto (2007) An R Package for Prototype Programming. http://cran.r-project.org/web/packages/proto/. Accessed 2 Oct 2008
Acknowledgment
This work is supported in part by the Austrian Grid Project of the Austrian Ministry of Education, Science and Culture (contract no. GZ 4003/2-VI/4c/2004).
Conflict of interest
The authors declare no conflict of interests. None of the authors is author or co-author of any tool that was discussed and evaluated in this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Knapp, B., Omasits, U., Frantal, S. et al. A critical cross-validation of high throughput structural binding prediction methods for pMHC. J Comput Aided Mol Des 23, 301–307 (2009). https://doi.org/10.1007/s10822-009-9259-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-009-9259-2