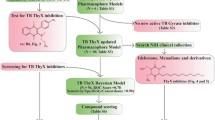
Abstract
Thymidine monophosphate kinase (TMPKmt) is an essential enzyme for nucleotide metabolism in Mycobacterium tuberculosis, and thus an attractive target for novel antituberculosis agents. In this work, we have explored the chemical space around the 2′,3′-bicyclic thymidine nucleus by designing and in silico screening of a virtual focused library selected via structure based methods to identify more potent analogs endowed with favorable ADME-related properties. In all the library members we have exchanged the ribose ring of the template with a cyclopentane moiety that is less prone to enzymatic degradation. In addition, we have replaced the six-membered 2′,3′-ring by a number of five-membered and six-membered heterocyclic rings containing alternative proton donor and acceptor groups, to exploit the interaction with the carboxylate groups of Asp9 and Asp163 as well as with several cationic residues present in the vicinity of the TMPKmt binding site. The three-dimensional structure of the TMPKmt complexed with 5-hydroxymethyl-dUMP, an analog of dTMP, was employed to develop a QSAR model, to parameterize a scoring function specific for the TMPKmt target and to select analogues which display the highest predicted binding to the target. As a result, we identified a small highly focused combinatorial subset of bicyclic thymidine analogues as virtual hits that are predicted to inhibit the mycobacterial TMPK in the submicromolar concentration range and to display favorable ADME-related properties.
Graphical abstract
















Similar content being viewed by others
Abbreviations
- ADME:
-
Adsorption, distribution, metabolism and excretion
- AZTMP:
-
3′-azido-2′-deoxythymidine monophosphate
- CFF91:
-
Consistent class II force field
- dTTP:
-
Deoxythymidine triphosphate
- dUMP:
-
Deoxyuridine monophosphate
- HB:
-
Hydrogen bond
- TEM:
-
5′-CH2OH dUMP inhibitor
- TMPKh :
-
Human thymidine monophosphate kinase
- TMPKmt :
-
Mycobacterium tuberculosis thymidine monophosphate kinase
References
Global Tuberculosis Control—Epidemiology, Strategy, Financing (2009) WHO report 2009.411, World Health Organization, Geneva. Retrieved from http://www.who.int/tb/publications/global_report/2009/en/index.html. 1/9/2010
Ginsberg AM, Spigelman M (2007) Challenges in tuberculosis drug research and development. Nat Med 13:290–294
WHO Global Task Force: Outlines Measures to Combat XDR-TB Worldwide (2006) WHO press release, World Health Organization, Geneva. Retrieved from http://www.who.int/mediacentre/news/notes/2006/np29/en/index.html. 1/9/2010
Mitnick CD, McGee B, Peloquin CA (2009) Tuberculosis pharmacotherapy: strategies to optimize patient care. Expert Opin Pharmacother 10:381–401
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry C III, Tekaia CF, Badcock K, Basham D, Brown D, Chillingworth DT, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544
Long MC, Parker WB (2006) Structure-activity relationship for nucleoside analogs as inhibitors or substrates of adenosine kinase from Mycobacterium tuberculosis. I. Modifications to the adenine moiety. Biochem Pharmacol 71:1671–1682
Williams KJ, Duncan K (2007) Current strategies for identifying and validating targets for new treatment-shortening drugs for TB. Curr Mol Med 7:297–307
Munier-Lehmann H, Chafotte A, Pochet S, Labesse G (2001) Thymidylate kinase of Mycobacterium tuberculosis: a chimera sharing properties common to eukaryotic and bacterial enzymes. Protein Sci 10:1195–1205
Anderson E (1973) In: Boyer PD (ed) The enzymes, vol 8. Academic Press, New York, pp 49–96
Sclafani RA, Fangman WL (1984) Yeast gene CDC8 encodes thymidylate kinase and is complemented by herpes thymidine kinase gene TK. Proc Natl Acad Sci USA 81:5821–5825
Li de la Sierra I, Munier-Lehmann H, Gilles AM, Bârzu O, Delarue M (2001) X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 A resolution. J Mol Biol 311:87–100
Li de la Sierra I, Munier-Lehmann H, Gilles AM, Bârzu O, Delarue M (2000) Crystallization and preliminary X-ray analysis of the thymidylate kinase from Mycobacterium tuberculosis. Acta Crystallogr Sect D Biol Crystallogr 56:226–228
Vanheusden V, Munier-Lehmann H, Pochet S, Herdewijna P, Van Calenbergh S (2002) Synthesis and evaluation of thymidine-5′-O-monophosphate analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Bioorg Med Chem Lett 12:2695–2698
Vanheusden V, Munier-Lehmann H, Froeyen M, Busson R, Rozenski J, Herdewijn P, Van Calenbergh S (2004) Discovery of bicyclic thymidine analogues as selective and high-affinity inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J Med Chem 47:6187–6194
Haouz A, Vanheusden V, Munier-Lehmann H, Froeyen M, Herdewijn P, Van Calenbergh S, Delarue M (2003) Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism. J Biol Chem 278:4963–4971
Pochet S, Dugué L, Douguet D, Labesse G, Munier-Lehmann H (2002) Nucleoside analogues as inhibitors of thymidylate kinases: possible therapeutic applications. ChemBioChem 3:108–110
Vanheusden V, Van Rompaey P, Munier-Lehmann H, Pochet S, Herdewijn P, Van Calenbergh S (2003) Thymidine and thymidine-5′-O-monophosphate analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Bioorg Med Chem Lett 13:3045–3048
Vanheusden V, Munier-Lehmann H, Froeyen M, Dugué L, Heyerick A, De Keukeleire D, Pochet S, Busson R, Herdewijn P, Van Calenbergh S (2003) 3′-C-branched-chain-substituted nucleosides and nucleotides as potent inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J Med Chem 46:3811–3821
Pochet S, Dugué L, Labesse G, Delepierre M, Munier-Lehmann H (2003) Comparative study of purine and pyrimidine nucleoside analogues acting on the thymidylate kinases of Mycobacterium tuberculosis and of humans. ChemBioChem 4:742–747
Van Daele I, Munier-Lehmann H, Hendrickx PM, Marchal G, Chavarot P, Froeyen M, Qing L, Martins CJ, Van Calenbergh S (2006) Synthesis and biological evaluation of bicyclic nucleosides as inhibitors of M. tuberculosis thymidylate kinase. ChemMedChem 1:1081–1090
Van Daele I, Munier-Lehmann H, Froeyen M, Balzarini J, Van Calenbergh S (2007) Rational design of 5′-thiourea-substituted alpha-thymidine analogues as thymidine monophosphate kinase inhibitors capable of inhibiting mycobacterial growth. J Med Chem 50:5281–5292
Gasse C, Douguet D, Huteau V, Marchal G, Munier-Lehmann H, Pochet S (2008) Substituted benzyl-pyrimidines targeting thymidine monophosphate kinase of Mycobacterium tuberculosis: synthesis and in vitro anti-mycobacterial activity. Bioorg Med Chem 16:6075–6085
Familiar O, Munier-Lehmann H, Negri A, Gago F, Douguet D, Rigouts L, Hernández A-I, Camarasa M-J, Pérez-Pérez M-J (2008) Exploring acyclic nucleoside analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. ChemMedChem 3:1083–1093
Frecer V, Burello E, Miertus S (2005) Combinatorial design of nonsymmetrical cyclic urea inhibitors of aspartic protease of HIV-1. Bioorg Med Chem 13:5492–5501
Frecer V, Megnassan E, Miertus S (2009) Design and in silico screening of combinatorial library of antimalarial analogs of triclosan inhibiting Plasmodium falciparum enoyl-acyl carrier protein reductase. Eur J Med Chem 44:3009–3019
Rungrotmongkol T, Frecer V, De-Eknamkul W, Hannongbua S, Miertus S (2009) Design of oseltamivir analogs inhibiting neuraminidase of avian influenza virus H5N1. Antivir Res 82:51–58
Rungrotmongkol T, Udommaneethanakit T, Frecer V, Miertus S (2010) Combinatorial design of avian influenza neuraminidase inhibitors containing pyrrolidine core with a reduced susceptibility to viral drug resistance. Comb Chem High Throughput Screen 13:268–277
Frecer V, Miertus S (2010) Design, structure-based focusing and in silico screening of combinatorial library of peptidomimetic inhibitors of Dengue virus NS2B-NS3 protease. J Comput Aided Mol Des 24:195–212
Pérez-Pérez M-J, Priego EM, Hernández AI, Camarasa MJ, Balzarini J, Liekens S (2005) Thymidine phosphorylase inhibitors: recent developments and potential therapeutic applications. Mini-Rev Med Chem 5:1113–1123
Kifli N, De Clercq E, Balzarini J, Simons C (2004) Novel bicyclic sugar modified nucleosides: synthesis, conformational analysis and antiviral evaluation. Bioorg Med Chem 12:3247–4252
Cerius2 Life Sciences software, version 4.6 (2002) Accelrys, San Diego, CA
Peters KP, Fauck J, Frommel C (1996) The automatic search for ligand binding sites in proteins of known three-dimensional structure using only geometric criteria. J Mol Biol 256:201–213
Jarvis RA, Patrick EA (1973) Clustering using a similarity measure based on shared near neighbors. IEEE Trans Comput C22:1025–1034
Seifert MH (2009) Targeted scoring functions for virtual screening. Drug Discov Today 14:562–569
Böhm HJ (1994) The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J Comput Aided Mol Des 8:243–256
Böhm HJ (1994) On the use of LUDI to search the Fine Chemicals Directory for ligands of proteins of known three-dimensional structure. J Comput Aided Mol Des 8:623–632
Muegge I, Martin YC (1999) A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J Med Chem 42:791–804
Muegge I, Martin YC, Hajduk PJ, Fesik SW (1999) Evaluation of PMF scoring in docking weak ligands to the FK506 binding protein. J Med Chem 42:2498–2503
Muegge I (2001) Effect of Ligand Volume Correction on PMF Scoring. J Comput Chem 22:418–425
Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Fogel LJ, Freer ST (1995) Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: conformationally flexible docking by evolutionary programming. Chem Biol 2:317–324
Verkhivker GM, Bouzida D, Gehlhaar DK, Reijto PA, Arthurs S, Colson AB, Freer ST, Larson V, Luty BA, Marrone T, Rose PW (2000) Deciphering common failures in molecular docking of ligand-protein complexes. J Comput Aided Mol Des 14:731–751
Holloway MK, McGaughey GB, Coburn CA, Stachel SJ, Jones KG, Stanton EL, Gregro AR, Lai M-T, Crouthamel M-C, Pietrak BL, Munshi SK (2007) Evaluating scoring functions for docking and designing beta-secretase inhibitors. Bioorg Med Chem Lett 17:823–827
Kontoyianni M, Sokol GS, McClellan LM (2005) Evaluation of library ranking efficacy in virtual screening. J Comput Chem 26:11–22
Krovat EM, Langer T (2004) Impact of scoring functions on enrichment in docking-based virtual screening: an application study on renin inhibitors. J Chem Inf Comput Sci 44:1123–1129
Wang R, Lu Y, Wang S (2003) Comparative evaluation of 11 scoring functions for molecular docking. J Med Chem 47:2287–2303
Insight-II Life Sciences software, version (2005) Accelrys, San Diego, CA
Frecer V, Berti F, Benedetti F, Miertus S (2008) Design of peptidomimetic inhibitors of aspartic protease of HIV-1 including–PheΨPro–core and favorable ADME properties. J Mol Graphics Model 27:376–387
Maple JR, Hwang MJ, Stockfish TP, Dinur U, Waldman M, Ewing CS, Hagler AT (1994) Derivation of class II force fields. I. Methodology and quantum force field for the alkyl functional group and alkane molecules. J Comput Chem 15:162–182
QikProp ADME Prediction software, version 3.2, Schrödinger, New York, NY
Duffy EM, Jorgensen WL (2000) Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J Am Chem Soc 122:2878–2888
Jorgensen WL, Duffy EM (2000) Prediction of drug solubility from Monte Carlo simulations. Bioorg Med Chem Lett 10:1155–1158
Jorgensen WL, Duffy EM (2002) Prediction of drug solubility from structure. Adv Drug Deliv Rev 54:355–366
Rappé AK, Goddard WA III (1991) Charge equilibration for molecular dynamics simulations. J Phys Chem 95:3358–3363
Available Chemicals Directory, version 3.0, Symyx Technologies, Santa Clara, CA
Blondin C, Serina L, Wiesmüller L, Gilles AM, Bârzu O (1994) Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal Biochem 220:219–222
Srinivasan J, Miller J, Kollman PA, Case DA (1998) Continuum solvent studies of the stability of RNA hairpin loops and helices. J Biol Struct Dyn 16:671–682
Chong LT, Duan Y, Wang L, Massova I, Kollman PA (1999) Molecular dynamics and free-energy calculations applied to affinity maturation in antibody 48G7. Proc Natl Acad Sci USA 96:14330–14335
Lee MR, Duan Y, Kollman PA (2000) Use of MM-PBSA in estimating the free energies of proteins: application to native, intermediates, nand unfolded villin headpiece. Proteins Struct Funct Genet 39:309–316
Wang J, Morin P, Wang W, Kollman PA (2001) Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc 123:5221–5230
Gilson MK, Honig B (1991) The inclusion of electrostatic hydration energies in molecular mechanics calculations. J Comput Aided Mol Des 5:5–20
Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrer A, Honig B (2002) Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem 23:128–137
Fischer S, Smith JC, Verma C (2001) Dissecting the vibrational entropy change on protein/ligand binding: burial of a water molecule in bovine pancreatic trypsin inhibitor. J Phys Chem B 105:8050–8055
Schwarzl SM, Tschopp TB, Smith JC, Fischer S (2002) Can the calculation of ligand binding free energies be improved with continuum solvent electrostatics and an ideal-gas entropy correction? J Comput Chem 23:1143–1149
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev 46:3–26
Acknowledgments
This work has been done within the ICS-UNIDO global program on Rational Drug Design and Discovery. Overall support of this work by ICS-UNIDO is gratefully acknowledged. We thank to Dr. Eugene Megnassan for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Frecer, V., Seneci, P. & Miertus, S. Computer-assisted combinatorial design of bicyclic thymidine analogs as inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J Comput Aided Mol Des 25, 31–49 (2011). https://doi.org/10.1007/s10822-010-9399-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-010-9399-4