Abstract
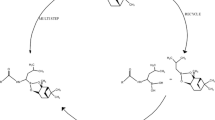
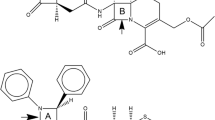
AmpC β-lactamase is a hydrolytic enzyme conferring resistance to β-lactam antibiotics in multiple Gram-negative bacteria. Therefore, identification of non-β-lactam compounds able to inhibit the enzyme is crucial for the development of novel antibacterial therapies. In general, AmpC inhibitors have to engage the highly solvent-exposed catalytic site of the enzyme. Therefore, understanding the implications of ligand–protein induced-fit and water-mediated interactions behind the inhibitor-enzyme recognition process is fundamental for undertaking structure-based drug design process. Here, we focus on boronic acids, a promising class of beta-lactamase covalent inhibitors. First, we optimized a docking protocol able to reproduce the experimentally determined binding mode of AmpC inhibitors bearing a boronic group. This goal was pursued (1) performing rigid and flexible docking calculations aiming to establish the role of the side chain conformations; and (2) investigating the role of specific water molecules in shaping the enzyme active site and mediating ligand protein interactions. Our calculations showed that some water molecules, conserved in the majority of the considered X-ray structures, are needed to correctly predict the binding pose of known covalent AmpC inhibitors. On this basis, we formalized our findings in a docking and scoring protocol that could be useful for the structure-based design of new boronic acid AmpC inhibitors.






Similar content being viewed by others
References
Drawz SM, Bonomo RA (2010) Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23(20065329):160–201
Drawz SM, Papp-Wallace KM, Bonomo RA (2014) New beta-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846
Farina D, Spyrakis F, Venturelli A, Cross S, Tondi D, Costi MP (2014) The inhibition of extended spectrum β-lactamases: hits and leads. Curr Med Chem 21:1405–1434
Ambler RP (1980) The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 289:321–331
Sgrignani J, Magistrato A, Dal Peraro M, Vila AJ, Carloni P, Pierattelli R (2012) On the active site of mononuclear B1 metallo-lactamases: a computational study. J Comput Aided Mol Des 26:425–435
Dal Peraro M, Carloni P (2010) Catalytic mechanism of metallo β-lactamases: insights from calculations and experiments. In: Matta F (ed) Quantum biochemistry. Wiley-VCH Verlag GmbH & Co. KGaA, New York, pp 605–622
Page MI, Badarau A (2008) The mechanisms of catalysis by metallo beta-lactamases. Bioinorg Chem Appl. doi:10.1155/2008/576297
Simona F, Magistrato A, Dal Peraro M, Cavalli A, Vila AJ, Carloni P (2009) Common mechanistic features among metallo-beta-lactamases: a computational study of Aeromonas hydrophila CphA enzyme. J Biol Chem 284:28164–28171
Dal Peraro M, Vila AJ, Carloni P, Klein ML (2007) Role of zinc content on the catalytic efficiency of B1 metallo beta-lactamases. J Am Chem Soc 129:2808–2816
Kiener PA, Waley SG (1978) Reversible inhibitors of penicillinases. Biochem J 169:197–204
Goldstein EJ, Citron DM, Tyrrell KL, Merriam CV (2013) In vitro activity of Biapenem plus RPX7009, a carbapenem combined with a serine beta-lactamase inhibitor, against anaerobic bacteria. Antimicrob Agents Chemother 57:2620–2630
Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931
Coupez B, Lewis RA (2006) Docking and scoring—theoretically easy, practically impossible? Curr Med Chem 13:2995–3003
Weill N, Therrien E, Campagna-Slater V, Moitessier N (2013) Methods for docking small molecules to macromolecules: a user’s perspective. 1. Theory Curr Pharm Des 20:3338–3359
Ouyang X, Zhou S, Ge Z, Li R, Kwoh CK (2013) CovalentDock Cloud: a web server for automated covalent docking. Nucl Acids Res 41(Web Server issue):W329–W332. doi:10.1093/nar/gkt406
Zhu K, Borrelli KW, Greenwood JR, Day T, Abel R, Farid RS, Harder E (2014) Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J Chem Inf Model 54:1932–1940
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748
Jones G, Willett P, Glen RC (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245:43–53
Sgrignani J, Bonaccini C, Grazioso G, Chioccioli M, Cavalli A, Gratteri P (2009) Insights into docking and scoring neuronal alpha4beta2 nicotinic receptor agonists using molecular dynamics simulations and QM/MM calculations. J Comput Chem 30:2443–2454
Lexa KW, Carlson HA (2012) Protein flexibility in docking and surface mapping. Q Rev Biophys 45:301–343
Roberts BC, Mancera RL (2008) Ligand-protein docking with water molecules. J Chem Inf Model 48:397–408
Kumar A, Zhang KYJ (2013) Investigation on the effect of key water molecules on docking performance in CSARdock exercise. J Chem Inf Model 53:1880–1892
London N, Miller RM, Krishnan S, Uchida K, Irwin JJ, Eidam O, Gibold L, Cimermančič P, Bonnet R, Shoichet BK, Taunton J (2014) Covalent docking of large libraries for the discovery of chemical probes. Nat Chem Biol 10:1066–1072
Knight JDR, Hamelberg D, McCammon JA, Kothary R (2009) The role of conserved water molecules in the catalytic domain of protein kinases. Proteins 76:527–535
Powers RA, Blazquez J, Weston GS, Morosini MI, Baquero F, Shoichet BK (1999) The complexed structure and antimicrobial activity of a non-beta-lactam inhibitor of AmpC beta-lactamase. Protein Sci 8:2330–2337
Caselli E, Powers RA, Blasczcak LC, Wu CY, Prati F, Shoichet BK (2001) Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases. Chem Biol 8:17–31
Tondi D, Powers RA, Caselli E, Negri MC, Blázquez J, Costi MP, Shoichet BK (2001) Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase. Chem Biol 8:593–611
Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK (2001) Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207–9214
Powers RA, Shoichet BK (2002) Structure-based approach for binding site identification on AmpC beta-lactamase. J Med Chem 45:3222–3234
Chen Y, Minasov G, Roth TA, Prati F, Shoichet BK (2006) The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution. J Am Chem Soc 128:2970–2976
Morandi S, Morandi F, Caselli E, Shoichet BK, Prati F (2008) Structure-based optimization of cephalothin-analogue boronic acids as beta-lactamase inhibitors. Bioorg Med Chem 16:1195–1205
Tondi D, Calo S, Shoichet BK, Costi MP (2010) Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors. Bioorg Med Chem Lett 20:3416–3419
Eidam O, Romagnoli C, Caselli E, Babaoglu K, Pohlhaus DT, Karpiak J, Bonnet R, Shoichet BK, Prati F (2010) Design, synthesis, crystal structures, and antimicrobial activity of sulfonamide boronic acids as beta-lactamase inhibitors. J Med Chem 53:7852–7863
Eidam O, Romagnoli C, Dalmasso G, Barelier S, Caselli E, Bonnet R, Shoichet BK, Prati F (2012) Fragment-guided design of subnanomolar beta-lactamase inhibitors active in vivo. Proc Natl Acad Sci USA 109:17448–17453
Frisch MJ, Trucks GW, Schlegel HB, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JJA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, revision A.02. Gaussian Inc., Wallingford, CT
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38:3098
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comp Chem 30:2785–2791
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comp Chem 31:455–461
Ouyang X, Zhou S, Su CTT, Ge Z, Li R, Kwoh CK (2013) CovalentDock: automated covalent docking with parameterized covalent linkage energy estimation and molecular geometry constraints. J Comp Chem 34:326–336
Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW (2007) Diverse, high-quality test set for the validation of protein–ligand docking performance. J Med Chem 50:726–741
Clark M, Cramer RD, Van Opdenbosch N (1989) Validation of the general purpose tripos 5.2 force field. J Comp Chem 10:982–1012
Verdonk ML, Chessari G, Cole JC, Hartshorn MJ, Murray CW, Nissink JW, Taylor RD, Taylor R (2005) Modeling water molecules in protein–ligand docking using GOLD. J Med Chem 48:6504–6515
Case DA, Darden TA, Cheatham TEIII, Simmerling CL, Wang J, Duke RE, Luo R, Walker C, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2012) Amber 12. University of California, San Francisco
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
For occupancy definition see http://www.ks.uiuc.edu/Research/vmd/plugins/volmapgui
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR (1984) J Chem Phys 81:3684
de Beer SBA, Vermeulen NPE, Oostenbrink C (2010) The role of water molecules in computational drug design. Curr Top Med Chem 10:55–66
De Vivo M (2011) Bridging quantum mechanics and structure-based drug design. Front Biosci 16:1619–1633
Sgrignani J, Magistrato A (2013) First-principles modeling of biological systems and structure-based drug-design. Curr Comput Aided Drug Des 9:15–34
Zhu K, Borrelli KW, Greenwood JR, Day T, Abel R, Farid RS, Harder E (2014) Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J Chem Inf Mod 54:1932–1940
Toledo Warshaviak D, Golan G, Borrelli KW, Zhu K, Kalid O (2014) Structure-based virtual screening approach for discovery of covalently bound ligands. J Chem Inf Mod 54:1941–1950
Acknowledgments
We acknowledge the CINECA and the Regione Lombardia award under the LISA initiative, for the availability of high performance computing resources and support. Financial support was obtained by the Italian Ministry of Education and Research through the Flagship (PB05) “InterOmics”.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sgrignani, J., Novati, B., Colombo, G. et al. Covalent docking of selected boron-based serine beta-lactamase inhibitors. J Comput Aided Mol Des 29, 441–450 (2015). https://doi.org/10.1007/s10822-015-9834-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-015-9834-7