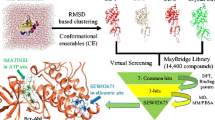
Abstract
Check point kinase 1 (Chk1) is an important protein in G2 phase checkpoint arrest required by cancer cells to maintain cell cycle and to prevent cell death. Therefore, Chk1 inhibitors should have potential as anti-cancer therapeutics. Docking-based comparative intermolecular contacts analysis (dbCICA) is a new three-dimensional quantitative structure activity relationship method that depends on the quality and number of contact points between docked ligands and binding pocket amino acid residues. In this presented work we implemented a novel combination of k-nearest neighbor/genetic function algorithm modeling coupled with dbCICA to select critical ligand-Chk1 contacts capable of explaining anti-Chk1 bioactivity among a long list of inhibitors. The finest set of contacts were translated into two valid pharmacophore hypotheses that were used as 3D search queries to screen the National Cancer Institute’s structural database for new Chk1 inhibitors. Three potent Chk1 inhibitors were discovered with IC50 values ranging from 2.4 to 69.7 µM.










Similar content being viewed by others
References
Carrassa L, Damia G (2011) Unleashing Chk1 in cancer therapy. Cell Cycle 10:2121–2128. doi:10.4161/cc.10.13.16398
Wang GT, Li G, Mantei RA, Chen Z, Kovar P, Gu W, Xiao Z, Zhang H, Sham HL, Sowin T, Rosenberg SH, Lin NH (2005) 1-(5-Chloro-2-alkoxyphenyl)-3-(5-cyanopyrazin-2-yl)ureas [correction of cyanopyrazi] as potent and selective inhibitors of Chk1 kinase: synthesis, preliminary SAR, and biological activities. J Med Chem 48:3118–3121. doi:10.1021/jm048989d
Zhou BB, Bartek J (2004) Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer 4:216–225. doi:10.1038/nrc1296
Hirose Y, Berger MS, Pieper RO (2001) Abrogation of the Chk1-mediated G(2) checkpoint pathway potentiates temozolomide-induced toxicity in a p53-independent manner in human glioblastoma cells. Cancer Res 61:5843–5849
Powell SN, DeFrank JS, Connell P, Eogan M, Preffer F, Dombkowski D, Tang W, Friend S (1995) Differential sensitivity of p53(−) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res 55:1643–1648
Kawabe T (2004) G2 checkpoint abrogators as anticancer drugs. Mol Cancer Ther 3:513–519
Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ (2001) Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene 20:7453–7463. doi:10.1038/sj.onc.1204942
Wang H, Wang X, Zhou XY, Chen DJ, Li GC, Iliakis G, Wang Y (2002) Ku affects the ataxia and Rad 3-related/CHK1-dependent S phase checkpoint response after camptothecin treatment. Cancer Res 62:2483–2487
Chen Z, Xiao Z, Chen J, Ng SC, Sowin T, Sham H, Rosenberg S, Fesik S, Zhang H (2003) Human Chk1 expression is dispensable for somatic cell death and critical for sustaining G2 DNA damage checkpoint. Mol Cancer Ther 2(6):543–548
Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, Piwnica-Worms H (2000) The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem 275:5600–5605. doi:10.1074/jbc.275.8.5600
Taha MO, Habash M, Al-Hadidi Z, Al-Bakri A, Younis K, Sisan S (2011) Docking-based comparative intermolecular contacts analysis as new 3-D QSAR concept for validating docking studies and in silico screening: NMT and GP inhibitors as case studies. J Chem Inf Model 51:647–669. doi:10.1021/ci100368t
Taha MO, Habash M, Khanfar MA (2014) The use of docking-based comparative intermolecular contacts analysis to identify optimal docking conditions within glucokinase and to discover of new GK activators. J Comput Aided Mol Des 28:509–547. doi:10.1007/s10822-014-9740-4
Al-Sha’er MA, Taha MO (2012) Application of docking-based comparative intermolecular contacts analysis to validate Hsp90alpha docking studies and subsequent in silico screening for inhibitors. J Mol Model 18:4843–4863. doi:10.1007/s00894-012-1479-z
Sharaf MA, Illman DL, Kowalski BR (1986) Chemometrics. Wiley, New York
Zheng W, Tropsha A (2000) Novel variable selection quantitative structure—property relationship approach based on the k-nearest-neighbor principle. J Chem Inf Comput Sci 40:185–194. doi:10.1021/ci980033m
Khanfar MA, Taha MO (2013) Elaborate ligand-based modeling coupled with multiple linear regression and k nearest neighbor QSAR analyses unveiled new nanomolar mTOR inhibitors. J Chem Inf Model 53(10):2587–2612. doi:10.1021/ci4003798
Tao ZF, Chen Z, Bui MH, Kovar P, Johnson E, Bouska J, Zhang H, Rosenberg S, Sowin T, Lin NH (2007) Macrocyclic ureas as potent and selective Chk1 inhibitors: an improved synthesis, kinome profiling, structure-activity relationships, and preliminary pharmacokinetics. Bioorg Med Chem Lett 17(23):6593–6601. doi:10.1016/j.bmcl.2007.09.063
Tao ZF, Li G, Tong Y, Stewart KD, Chen Z, Bui MH, Merta P, Park C, Kovar P, Zhang H, Sham HL, Rosenberg SH, Sowin TJ, Lin NH (2007) Discovery of 4′-(1,4-dihydro-indeno[1,2-c]pyrazol-3-yl)-benzonitriles and 4′-(1,4-dihydro-indeno[1,2-c]pyrazol-3-yl)-pyridine-2′-carbonitriles as potent checkpoint kinase 1 (Chk1) inhibitors. Bioorg Med Chem Lett 17(21):5944–5951. doi:10.1016/j.bmcl.2007.07.102
Tao ZF, Li G, Tong Y, Chen Z, Merta P, Kovar P, Zhang H, Rosenberg SH, Sham HL, Sowin TJ, Lin NH (2007) Synthesis and biological evaluation of 4′-(6,7-disubstituted-2,4-dihydro-indeno[1,2-c]pyrazol-3-yl)-biphenyl-4-ol as potent Chk1 inhibitors. Bioorg Med Chem Lett 17(15):4308–4315. doi:10.1016/j.bmcl.2007.05.027
Tao ZF, Wang L, Stewart KD, Chen Z, Gu W, Bui MH, Merta P, Zhang H, Kovar P, Johnson E, Park C, Judge R, Rosenberg S, Sowin T, Lin NH (2007) Structure-based design, synthesis, and biological evaluation of potent and selective macrocyclic checkpoint kinase 1 inhibitors. J Med Chem 50(7):1514–1527. doi:10.1021/jm061247v
Venkatachalam CM, Jiang X, Oldfield T, Waldman M (2003) LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model 21:289–307. doi:10.1016/S1093-3263(02)00164-X
Rogers D, Hopfinger AJ (1994) Application of genetic function approximation to quantitative structure-Activity-relationships and quantitative structure-property relationships. J Chem Inf Comput Sci 34(4):854–866. doi:10.1021/Ci00020a020
Verdonk ML, Berdini V, Hartshorn MJ, Mooij WT, Murray CW, Taylor RD, Watson P (2004) Virtual screening using protein-ligand docking: avoiding artificial enrichment. J Chem Inf Comput Sci 44(3):793–806. doi:10.1021/ci034289q
Abu Hammad AM, Afifi FU, Taha MO (2007) Combining docking, scoring and molecular field analyses to probe influenza neuraminidase-ligand interactions. J Mol Graph Model 26(2):443–456. doi:10.1016/j.jmgm.2007.02.002
Abu-Hammad A, Zalloum WA, Zalloum H, Abu-Sheikha G, Taha MO (2009) Homology modeling of MCH1 receptor and validation by docking/scoring and protein-aligned CoMFA. Eur J Med Chem 44(6):2583–2596. doi:10.1016/j.ejmech.2009.01.031
Taha MO, AlDamen MA (2005) Effects of variable docking conditions and scoring functions on corresponding protein-aligned comparative molecular field analysis models constructed from diverse human protein tyrosine phosphatase 1B inhibitors. J Med Chem 48(25):8016–8034. doi:10.1021/jm058047o
Homans SW (2007) Water, water everywhere–except where it matters? Drug Discov Today 12(13–14):534–539. doi:10.1016/j.drudis.2007.05.004
Song CM, Lim SJ, Tong JC (2009) Recent advances in computer-aided drug design. Brief Bioinform 10(5):579–591. doi:10.1093/bib/bbp023
Jorgensen WL (2009) Efficient drug lead discovery and optimization. Acc Chem Res 42(6):724–733. doi:10.1021/ar800236t
Leach AR, Shoichet BK, Peishoff CE (2006) Prediction of protein-ligand interactions. Docking and scoring: successes and gaps. J Med Chem 49(20):5851–5855. doi:10.1021/jm060999m
Krissinel E (2010) Crystal contacts as nature’s docking solutions. J Comput Chem 31(1):133–143. doi:10.1002/jcc.21303
Ni ZJ, Barsanti P, Brammeier N, Diebes A, Poon DJ, Ng S, Pecchi S, Pfister K, Renhowe PA, Ramurthy S, Wagman AS, Bussiere DE, Le V, Zhou Y, Jansen JM, Ma S, Gesner TG (2006) 4-(Aminoalkylamino)-3-benzimidazole-quinolinones as potent CHK-1 inhibitors. Bioorg Med Chem Lett 16(12):3121–3124. doi:10.1016/j.bmcl.2006.03.059
Foloppe N, Fisher LM, Howes R, Kierstan P, Potter A, Robertson AG, Surgenor AE (2005) Structure-based design of novel Chk1 inhibitors: insights into hydrogen bonding and protein-ligand affinity. J Med Chem 48(13):4332–4345. doi:10.1021/jm049022c
Triballeau N, Bertrand H-O, Acher F (2006) Are you sure you have a good model? In: Hoffmann RD (ed) Langer T. Pharmacophores and Pharmacophore Searches, Wiley, pp 325–364
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26. doi:10.1016/S0169-409X(00)00129-0
Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623. doi:10.1021/jm020017n
Reader JC, Matthews TP, Klair S, Cheung KM, Scanlon J, Proisy N, Addison G, Ellard J, Piton N, Taylor S, Cherry M, Fisher M, Boxall K, Burns S, Walton MI, Westwood IM, Hayes A, Eve P, Valenti M, de Haven Brandon A, Box G, van Montfort RL, Williams DH, Aherne GW, Raynaud FI, Eccles SA, Garrett MD, Collins I (2011) Structure-guided evolution of potent and selective CHK1 inhibitors through scaffold morphing. J Med Chem 54(24):8328–8342. doi:10.1021/jm2007326
Triballeau N, Acher F, Brabet I, Pin JP, Bertrand HO (2005) Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem 48(7):2534–2547. doi:10.1021/jm049092j
Kirchmair J, Markt P, Distinto S, Wolber G, Langer T (2008) Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection–what can we learn from earlier mistakes? J Comput Aided Mol Des 22(3–4):213–228. doi:10.1007/s10822-007-9163-6
Dudkin VY, Rickert K, Kreatsoulas C, Wang C, Arrington KL, Fraley ME, Hartman GD, Yan Y, Ikuta M, Stirdivant SM, Drakas RA, Walsh ES, Hamilton K, Buser CA, Lobell RB, Sepp-Lorenzino L (2012) Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates. Bioorg Med Chem Lett 22(7):2609–2612. doi:10.1016/j.bmcl.2012.01.110
Taha MO, Al-Sha’er MA, Khanfar MA, Al-Nadaf AH (2014) Discovery of nanomolar phosphoinositide 3-kinase gamma (PI3Kgamma) inhibitors using ligand-based modeling and virtual screening followed by in vitro analysis. Eur J Med Chem 84:454–465. doi:10.1016/j.ejmech.2014.07.056
Al-Sha’er MA, Khanfar MA, Taha MO (2014) Discovery of novel urokinase plasminogen activator (uPA) inhibitors using ligand-based modeling and virtual screening followed by in vitro analysis. J Mol Model 20(1):2080. doi:10.1007/s00894-014-2080-4
Abuhamdah S, Habash M, Taha MO (2013) Elaborate ligand-based modeling coupled with QSAR analysis and in silico screening reveal new potent acetylcholinesterase inhibitors. J Comput Aided Mol Des 27(12):1075–1092. doi:10.1007/s10822-013-9699-6
Matthews TP, Jones AM, Collins I (2013) Structure-based design, discovery and development of checkpoint kinase inhibitors as potential anticancer therapies. Expert Opin Drug Discov 8(6):621–640. doi:10.1517/17460441.2013.788496
Imbert P-E, Unterreiner V, Siebert D, Gubler H, Parker C, Gabriel D (2007) Recommendations for the reduction of compound artifacts in time-resolved fluorescence resonance energy transfer assays. Assay Drug Dev Technol 5(3):363–372. doi:10.1089/adt.2007.073
Acknowledgments
The authors thank the Deanship of Scientific Research and Hamdi-Mango Center for Scientific Research at the University of Jordan for their generous funds. The authors are also thankful to the National Cancer Institute for freely providing NCI hits. (Additional Supporting Information may be found in the online version of this article.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jaradat, N.J., Khanfar, M.A., Habash, M. et al. Combining docking-based comparative intermolecular contacts analysis and k-nearest neighbor correlation for the discovery of new check point kinase 1 inhibitors. J Comput Aided Mol Des 29, 561–581 (2015). https://doi.org/10.1007/s10822-015-9848-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-015-9848-1