Abstract
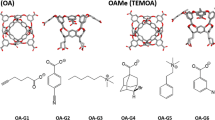
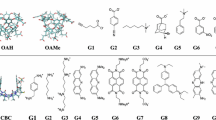
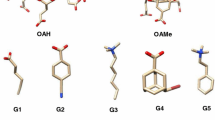
We review our performance in the SAMPL5 challenge for predicting host–guest binding affinities using the movable type (MT) method. The challenge included three hosts, acyclic Cucurbit[2]uril and two octa-acids with and without methylation at the entrance to their binding cavities. Each host was associated with 6–10 guest molecules. The MT method extrapolates local energy landscapes around particular molecular states and estimates the free energy by Monte Carlo integration over these landscapes. Two blind submissions pairing MT with variants of the KECSA potential function yielded mean unsigned errors of 1.26 and 1.53 kcal/mol for the non-methylated octa-acid, 2.83 and 3.06 kcal/mol for the methylated octa-acid, and 2.77 and 3.36 kcal/mol for Cucurbit[2]uril host. While our results are in reasonable agreement with experiment, we focused on particular cases in which our estimates gave incorrect results, particularly with regard to association between the octa-acids and an adamantane derivative. Working on the hypothesis that differential solvation effects play a role in effecting computed binding affinities for the parent octa-acid and the methylated octa-acid and that the ligands bind inside the pockets (rather than on the surface) we devised a new solvent accessible surface area term to better quantify solvation energy contributions in MT based studies. To further explore this issue a, molecular dynamics potential of mean force (PMF) study indicates that, as found by our docking calculations, the stable binding mode for this ligand is inside (rather than surface bound) the octa-acid cavity whether the entrance is methylated or not. The PMF studies also obtained the correct order for the methylation-induced change in binding affinities and associated the difference, to a large extent to differential solvation effects. Overall, the SAMPL5 challenge yielded in improvements our solvation modeling and also demonstrated the need for thorough validation of input data integrity prior to any computational analysis.







Similar content being viewed by others
References
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) J Comput Aided Mol Des 24(4):259
Guthrie JP (2009) J Phys Chem B 113(14):4501
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) J Comput Aided Mol Des 28(4):305
Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK (2012) J Comput Aided Mol Des 26(5):475
Skillman AG (2012) J Comput Aided Mol Des 26(5):473
Benson ML, Faver JC, Ucisik MN, Dashti DS, Zheng Z, Merz KM (2012) J Comput Aided Mol Des 26(5):647
Yin J, Henriksen NM, Slochower DR, Shirts MR, Chiu MW, Mobley DL, Gilson MK (2016) J Comput Aided Mol Des. doi:10.1007/s10822-016-9974-4
Chang CE, Gilson MK (2004) J Am Chem Soc 126(40):13156
Chen W, Chang CE, Gilson MK (2004) Biophys J 87(5):3035
Houk KN, Leach AG, Kim SP, Zhang XY (2003) Angew Chem Int Ed 42(40):4872
Liu SM, Ruspic C, Mukhopadhyay P, Chakrabarti S, Zavalij PY, Isaacs L (2005) J Am Chem Soc 127(45):15959
Gilberg L, Zhang B, Zavalij PY, Sindelar V, Isaacs L (2015) Org Biomol Chem 13(13):4041
Zhang B, Isaacs L (2014) J Med Chem 57(22):9554
Hettiarachchi G, Nguyen D, Wu J, Lucas D, Ma D, Isaacs L, Briken V (2010) PLoS One 5(5):e10514. doi:10.1371/journal.pone.0010514
Lagona J, Mukhopadhyay P, Chakrabarti S, Isaacs L (2005) Angew Chem Int Ed 44(31):4844
Rogers KE, Ortiz-Sanchez JM, Baron R, Fajer M, de Oliveira CAF, McCammon JA (2013) J Chem Theory Comput 9(1):46
Choudhury R, Gupta S, Da Silva JP, Ramamurthy V (2013) J Org Chem 78(5):1824
Porel M, Jayaraj N, Kaanumalle LS, Maddipatla MVSN, Parthasarathy A, Ramamurthy V (2009) Langmuir 25(6):3473
Gibb CLD, Gibb BC (2014) J Comput Aided Mol Des 28(4):319
Gibb CLD, Gibb BC (2004) J Am Chem Soc 126(37):11408
Liu SM, Whisenhunt-Ioup SE, Gibb CLD, Gibb BC (2011) Supramol Chem 23(6):480
Gan HY, Benjamin CJ, Gibb BC (2011) J Am Chem Soc 133(13):4770
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) J Comput Aided Mol Des 27(3):221
Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH (2011) J Chem Theory Comput 7(2):525
Rostkowski M, Olsson MHM, Sondergaard CR, Jensen JH (2011) BMC Struct Biol. doi:10.1186/1472-6807-11-6
Jorgensen WL, Tiradorives J (1988) J Am Chem Soc 110(6):1657
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) J Phys Chem B 105(28):6474
LigPrep (2015) Version 3.6. Schrödinger, LLC, New York
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) J Comput Aided Mol Des 21(12):681
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) J Med Chem 47(7):1739
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) J Med Chem 49(21):6177
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) J Med Chem 47(7):1750
Macromodel (2015) Schrödinger, LLC, New York
Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC (1990) J Comput Chem 11(4):440
Polak E, Ribiere G (1969) Rev Fr Inf Rech Oper 3(16):35
Kolossvary I, Guida WC (1996) J Am Chem Soc 118(21):5011
Zheng Z, Merz KM (2013) J Chem Inf Model 53(5):1073
Cleveland WS (1979) J Am Stat Assoc 74(368):829
Cleveland WS (1981) Am Stat 35(1):54
Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM (1992) J Comput Chem 13(8):1011
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25(9):1157
Berendsen HJC, Grigera JR, Straatsma TP (1987) J Phys Chem 91(24):6269
Zheng Z, Wang T, Li PF, Merz KM (2015) J Chem Theory Comput 11(2):667
Maestro (2015) Schrödinger, LLC, New York
Acknowledgments
We would like to acknowledge the SAMPL5 organizers for providing the data and platform for the blind challenge and global communication. NB would like to acknowledge Mr. Dario Gioia for numerous discussions related to docking of host–guest systems.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nupur Bansal and Zheng Zheng have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bansal, N., Zheng, Z., Cerutti, D.S. et al. On the fly estimation of host–guest binding free energies using the movable type method: participation in the SAMPL5 blind challenge. J Comput Aided Mol Des 31, 47–60 (2017). https://doi.org/10.1007/s10822-016-9980-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9980-6