Abstract
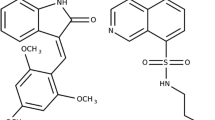
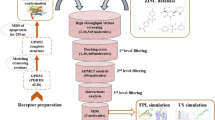
Despite a wealth of persuasive evidence for the involvement of human small C-terminal domain phosphatase 1 (Scp1) in the impairment of neuronal differentiation and in Huntington’s disease, small-molecule inhibitors of Scp1 have been rarely reported so far. This study aims to the discovery of both competitive and allosteric Scp1 inhibitors through the two-track virtual screening procedure. By virtue of the improvement of the scoring function by implementing a new molecular solvation energy term and by reoptimizing the atomic charges for the active-site Mg2+ ion cluster, we have been able to identify three allosteric and five competitive Scp1 inhibitors with low-micromolar inhibitory activity. Consistent with the results of kinetic studies on the inhibitory mechanisms, the allosteric inhibitors appear to be accommodated in the peripheral binding pocket through the hydrophobic interactions with the nonpolar residues whereas the competitive ones bind tightly in the active site with a direct coordination to the central Mg2+ ion. Some structural modifications to improve the biochemical potency of the newly identified inhibitors are proposed based on the binding modes estimated with docking simulations.









Similar content being viewed by others
References
Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP (2006) Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell 24:759–770
Kokabu S, Ohte S, Sasanuma H, Shin M, Yoneyama K, Murata E, Kanomata K, Nojima J, Ono Y, Yoda T, Fukuda T, Katagiri T (2011) Suppression of BMP-Smad signaling axis- induced osteoblastic differentiation by small C-terminal domain phosphatase 1, a Smad phosphatase. Mol Endocrinol 25:474–481
Nesti E, Corson GM, McCleskey M, Oyer JA, Mandel G (2014) C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc Natl Acad Sci USA 111:E3929–E3936
Sun T, Fu J, Shen T, Lin X, Liao L, Feng XH, Xu J (2016) The small C-terminal domain phosphatase 1 inhibits cancer cell migration and invasion by dephosphorylating Ser(P)68-Twist1 to accelerate Twist1 protein degradation. J Biol Chem 291:11518–11528
Visvanathan J, Lee S, Lee B, Lee JW, Lee SK (2007) The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21:744–749
Wang W, Liao P, Shen M, Chen T, Chen Y, Li Y, Lin X, Ge X, Wang P (2016) SCP1 regulates c-Myc stability and functions through dephosphorylating c-Myc Ser62. Oncogene 35:491–500
Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E (2003) Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 35:76–83
Palm K, Belluardo N, Metsis M, Timmusk T (1998) Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci 18:1280–1296
Zuccato C, Valenza M, Cattaneo E (2010) Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev 90:905–981
Aronin N, Chase K, Young C, Sapp E, Schwarz C, Matta N, Kornreich R, Landwehrmeyer B, Bird E, Beal MF, Vonsattel JP, Smith T, Carraway R, Boyce FM, Young AB, Penney JB, DiFiglia M (1995) CAG expansion affects the expression of mutant huntingtin in the Huntington’s disease brain. Neuron 15:1193–1201
Soldati C, Bithell A, Johnston C, Wong KY, Stanton LW, Buckley NJ (2013) Dysregulation of REST-regulated coding and non-coding RNAs in a cellular model of Huntington’s disease. J Neurochem 124:418–430
Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ (2008) SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452:370–374
Shi F, Yang Y, Wang T, Kouadir M, Zhao D, Hu S (2016) Cellular prion protein promotes neuronal differentiation of adipose-derived stem cells by upregulating miRNA-124. J Mol Neurosci 59:48–55
Liu T, Im W, Mook-Jung I, Kim M (2015) MicroRNA-124 slows down the progression of Huntington’s disease by promoting neurogenesis in the striatum. Neural Regen Res 10:786–791
Zhang M, Cho EJ, Burstein G, Siegel D, Zhang Y (2011) Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem Biol 6:511–519
Zilian D, Sotriffer CA (2013) SFCscoreRF: a random forest-based scoring function for improved affinity prediction of protein–ligand complexes. J Chem Inf Model 53:1923–1933
Huang SY, Grinter SZ, Zou X (2010) Scoring functions and their evaluation methods for protein-ligand docking: recent advances and future directions. Phys Chem Chem Phys 12:12899–12908
Kamenski T, Heilmeier S, Meinhart A, Cramer P (2004) Structure and mechanism of RNA polymerase II CTD phosphatases. Mol Cell 15:399–407
Bayly CA, Cieplak P, Cornell WD, Kollman PA (1993) A well behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97:10269–10280
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev 23:3–25
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Park H, Lee J, Lee S (2006) Critical assessment of the automated AutoDock as a new docking tool for virtual screening. Proteins 65:549–554
Mehler EL, Solmajer T (1991) Electrostatic effects in proteins: comparison of dielectric and charge models. Protein Eng 4:903–910
Stouten PFW, Frömmel C, Nakamura H, Sander C (1993) An effective solvation term based on atomic occupancies for use in protein simulations. Mol Simul 10:97–120
Chung KC, Park H (2015) Accuracy enhancement in the estimation of molecular hydration free energies by implementing the intramolecular hydrogen bond effects. J Cheminform 7:57
Shoichet BK, Leach AR, Kuntz ID (1999) Ligand solvation in molecular docking. Proteins 34:4–16
Fox T, Kollman PA (1998) Application of the RESP methodology in the parameterization of organic solvents. J Phys Chem B 102:8070–8079
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117:5179–5197
Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740
Allen KN, Dunaway-Mariano D (2004) Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem Sci 29:495–503
Lizunov AY, Gonchar AL, Zaitseva NI, Zosimov VV (2015) Accounting for intraligand interactions in flexible ligand docking with a PMF-based scoring function. J Chem Inf Model 55:2121–2137
Rafi SB, Hearn BR, Vedantham P, Jacobson MP, Renslo AR (2012) Predicting and improving the membrane permeability of peptidic small molecules. J Med Chem 55:3163–3169
Acknowledgements
This research was supported by BioNano Health-Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as Global Frontier Project (H-GUARD_2014M3A6B2060507) and by the Bio & Medical Technology Development Programs of the National Research Foundation, funded by the Korean Government (2015M3A9B5030308 and 2014M3A9B6070243).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, H., Lee, H.S., Ku, B. et al. Two-track virtual screening approach to identify both competitive and allosteric inhibitors of human small C-terminal domain phosphatase 1. J Comput Aided Mol Des 31, 743–753 (2017). https://doi.org/10.1007/s10822-017-0037-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0037-2