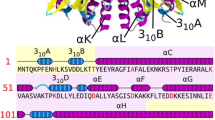
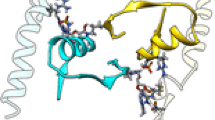
Abstract
I-DmoI, from the hyperthermophilic archaeon Desulfurococcus mobilis, belongs to the LAGLIDADG homing endonuclease protein family. Its members are highly specific enzymes capable of recognizing long DNA target sequences, thus providing potential tools for genome manipulation. Working towards this particular application, many efforts have been made to generate mesophilic variants of I-DmoI that function at lower temperatures than the wild-type. Here, we report a structural and computational analysis of two I-DmoI mesophilic mutants. Despite very limited structural variations between the crystal structures of these variants and the wild-type, a different dynamical behaviour near the cleavage sites is observed. In particular, both the dynamics of the water molecules and the protein perturbation effect on the cleavage site correlate well with the changes observed in the experimental enzymatic activity.








Similar content being viewed by others
References
Chan SH, Stoddard BL, Xu SY (2011) Natural and engineered nicking endonucleases—from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res 39:1–18. https://doi.org/10.1093/nar/gkq742
Galetto R, Duchateau P, Paques F (2009) Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin Biol Ther 9:1289–1303. https://doi.org/10.1517/14712590903213669
Molina R et al (2012) Non-specific protein–DNA interactions control I-CreI target binding and cleavage. Nucleic Acids Res 40:6936–6945. https://doi.org/10.1093/nar/gks320
Munoz IG et al (2011) Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res 39:729–743. https://doi.org/10.1093/nar/gkq801
Paques F, Duchateau P (2007) Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther 7:49–66
Stoddard BL (2005) Homing endonuclease structure and function. Q Rev Biophys 38:49–95. https://doi.org/10.1017/S0033583505004063
Marcaida MJ et al (2008) Crystal structure of I-DmoI in complex with its target DNA provides new insights into meganuclease engineering. Proc Natl Acad Sci USA 105:16888–16893. https://doi.org/10.1073/pnas.0804795105
Molina R et al (2016) Key players in I-DmoI endonuclease catalysis revealed from structure and dynamics. ACS Chem Biol 11:1401–1407. https://doi.org/10.1021/acschembio.5b00730
Molina R et al (2015) Engineering a nickase on the homing endonuclease I-DmoI scaffold. J Biol Chem 290:18534–18544. https://doi.org/10.1074/jbc.M115.658666
Dalgaard JZ, Garrett RA, Belfort M (1993) A site-specific endonuclease encoded by a typical archaeal intron. Proc Natl Acad Sci USA 90:5414–5417
Prieto J et al (2008) Generation and analysis of mesophilic variants of the thermostable archaeal I-DmoI homing endonuclease. J Biol Chem 283:4364–4374. https://doi.org/10.1074/jbc.M706323200
Amadei A, Linssen AB, Berendsen HJ (1993) Essential dynamics of proteins. Proteins 17:412–425. https://doi.org/10.1002/prot.340170408
Chevalier BS, Monnat RJ, Jr. & Stoddard BL (2001) The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat Struct Biol 8:312–316. https://doi.org/10.1038/86181
Dupureur CM (2008) Roles of metal ions in nucleases. Curr Opin Chem Biol 12:250–255. https://doi.org/10.1016/j.cbpa.2008.01.012
Dupureur CM (2010) One is enough: insights into the two-metal ion nuclease mechanism from global analysis and computational studies. Metallomics 2:609–620. https://doi.org/10.1039/c0mt00013b
Ivanov I, Tainer JA, McCammon JA (2007) Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc Natl Acad Sci USA 104:1465–1470. https://doi.org/10.1073/pnas.0603468104
Molina R et al (2015) Visualizing phosphodiester-bond hydrolysis by an endonuclease. Nat Struct Mol Biol 22:65–72. https://doi.org/10.1038/nsmb.2932
Aragones AC et al (2016) Electrostatic catalysis of a Diels-Alder reaction. Nature 531:88–91. https://doi.org/10.1038/nature16989
Amadei A, D’Alessandro M, Paci M, Di Nola A, Aschi M (2006) On the effect of a point mutation on the reactivity of CuZn superoxide dismutase: a theoretical study. J Phys Chem B 110:7538–7544. https://doi.org/10.1021/jp057095h
Shaik S, de Visser SP, Kumar D (2004) External electric field will control the selectivity of enzymatic-like bond activations. J Am Chem Soc 126:11746–11749. https://doi.org/10.1021/ja047432k
Arnould S et al (2006) Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol 355:443–458. https://doi.org/10.1016/j.jmb.2005.10.065
Epinat JC et al (2003) A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res 31:2952–2962
Redondo P, Prieto J, Ramos E, Blanco FJ, Montoya G (2007) Crystallization and preliminary X-ray diffraction analysis on the homing endonuclease I-Dmo-I in complex with its target DNA. Acta Crystallogr F 63:1017–1020. https://doi.org/10.1107/S1744309107049706
Kabsch W (2010) Xds. Acta Crystallogr D 66:125–132. https://doi.org/10.1107/S0907444909047337
Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D 62:72–82. https://doi.org/10.1107/S0907444905036693
McCoy AJ et al (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674. https://doi.org/10.1107/S0021889807021206
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D 66:486–501. https://doi.org/10.1107/S0907444910007493
Adams PD et al (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66:213–221. https://doi.org/10.1107/S0907444909052925
Hess B, Bekker H, Brendsen HJC, Fraaije J (1997) LINCS: a linear constant solver for molecular simulations. J Comput Chem 18:1463–1472
Darden T, Tork D, Pedersen L (1997) Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J Chem Phys 98:10089
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Hornak V et al (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725. https://doi.org/10.1002/prot.21123
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101. https://doi.org/10.1063/1.2408420
Luzar A, Chandler D Hydrogen-bond kinetics in liquid water. Nature 379:55–57. https://doi.org/10.1038/379055a0
Acknowledgements
This work was supported by Ministero dell’Istruzione, Università e Ricerca (R. Levi-Montalcini fellow to M.D.) and by Sapienza, University of Rome (Grant “Ateneo 2015”). We acknowledge CINECA Supercomputing Center, NVIDIA Academic Program, the Dept. of Chemistry for computational resources and the staffs at ALBA and SLS synchrotrons for helping in data collection.
Author information
Authors and Affiliations
Contributions
JA performed the Molecular Dynamics simulations; MJM performed the crystallization assays and X-ray data collection; MJM and RM carried out the crystal data processing, model building and refinement; RM, MJM, JP and GM were involved in the crystallographic analysis; J.A. and R.M. prepared the figures; JA and MD analysed the trajectories; RM and MD discussed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alba, J., Marcaida, M.J., Prieto, J. et al. Structure and dynamics of mesophilic variants from the homing endonuclease I-DmoI. J Comput Aided Mol Des 31, 1063–1072 (2017). https://doi.org/10.1007/s10822-017-0087-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0087-5