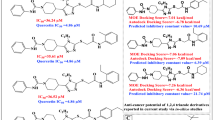
Abstract
Wee1 plays a critical role in the arrest of G2/M cell cycle for DNA repair before entering mitosis. Many cancer cells have been identified as overexpression of Wee1. In this research, pharmacophore modeling, molecular docking and molecular dynamics simulation approaches were constructed to identify novel potential Wee1 inhibitors. A compound 8 was found to have a novel skeleton against Wee1 with an IC50 value of 22.32 µM and a Ki value of 13.11 µM. Kinetic assays were employed to evaluate the compound 8 as a competitive inhibitor. Compound 8 was tested against A-549 tumor cell lines with IC50 value of 17.8 µM. To investigate the intermolecular interaction of Wee1 and compound 8, further molecular dynamics simulations were performed. It indicates that the binding mode of compound 8 and reference ligand is similar. The active core scaffold of compound 8 could represent a promising lead compound for studying Wee1 and be used for further structural optimization to design more potent Wee1 inhibitors.














Similar content being viewed by others
References
Nurse P, Thuriaux P (1980) Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics 96(3):627–637
Watanabe N, Broome M, Hunter T (1995) Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. Embo J 14(9):1878–1891
Mak JP, Man WY, Ma HT, Poon RY (2014) Pharmacological targeting the ATR-CHK1-WEE1 axis involves balancing cell growth stimulation and apoptosis. Oncotarget 5(21):10546–10557. https://doi.org/10.18632/oncotarget.2508
Den Haese GJ, Walworth N, Carr AM, Gould KL (1995) The Wee1 protein kinase regulates T14 phosphorylation of fission yeast Cdc2. Mol Biol Cell 6(4):371–385. https://doi.org/10.1091/mbc.6.4.371
Rowley R, Hudson J, Young PG (1992) The wee1 protein kinase is required for radiation-induced mitotic delay. Nature 356(6367):353–355. https://doi.org/10.1038/356353a0
De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJ, Wurdinger T (2011) WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res 17(13):4200–4207. https://doi.org/10.1158/1078-0432.ccr-10-2537
Mizuarai S, Yamanaka K, Itadani H, Arai T, Nishibata T, Hirai H, Kotani H (2009) Discovery of gene expression-based pharmacodynamic biomarker for a p53 context-specific anti-tumor drug Wee1 inhibitor. Mol Cancer 8(1):34. https://doi.org/10.1186/1476-4598-8-34
Van Linden AA, Baturin D, Ford JB, Fosmire SP, Gardner L, Korch C, Reigan P, Porter CC (2013) Inhibition of Wee1 sensitizes cancer cells to antimetabolite chemotherapeutics in vitro and in vivo, independent of p53 functionality. Mol Cancer Ther 12(12):2675–2684. https://doi.org/10.1158/1535-7163.MCT-13-0424
Pappano WN, Qian Z, Tucker LA, Tse C, Wang J (2014) Genetic inhibition of the atypical kinase Wee1 selectively drives apoptosis of p53 inactive tumor cells. BMC Cancer 14(1):430. https://doi.org/10.1186/1471-2407-14-430
Hashimoto O, Shinkawa M, Torimura T, Nakamura T, Selvendiran K, Sakamoto M, Koga H, Ueno T, Sata M (2006) Cell cycle regulation by the Wee1 inhibitor PD0166285, pyrido [2,3-d] pyimidine, in the B16 mouse melanoma cell line. BMC Cancer 6:292. https://doi.org/10.1186/1471-2407-6-292
Bridges KA, Hirai H, Buser CA, Brooks C, Liu H, Buchholz TA, Molkentine JM, Mason KA, Meyn RE (2011) MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin Cancer Res 17(17):5638–5648. https://doi.org/10.1158/1078-0432.ccr-11-0650
Palmer BD, Thompson AM, Booth RJ, Dobrusin EM, Kraker AJ, Lee HH, Lunney EA, Mitchell LH, Ortwine DF, Smaill JB, Swan LM, Denny WA (2006) 4-Phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-dione inhibitors of the checkpoint kinase Wee1. Structure-activity relationships for chromophore modification and phenyl ring substitution. J Med Chem 49(16):4896–4911. https://doi.org/10.1021/jm0512591
Pokorny JL, Calligaris D, Gupta SK, Iyekegbe DO Jr, Mueller D, Bakken KK, Carlson BL, Schroeder MA, Evans DL, Lou Z, Decker PA, Eckel-Passow JE, Pucci V, Ma B, Shumway SD, Elmquist WF, Agar NY, Sarkaria JN (2015) The efficacy of the wee1 inhibitor MK-1775 combined with temozolomide is limited by heterogeneous distribution across the blood-brain barrier in glioblastoma. Clin Cancer Res 21(8):1916–1924. https://doi.org/10.1158/1078-0432.ccr-14-2588
Rajeshkumar NV, De Oliveira E, Ottenhof N, Watters J, Brooks D, Demuth T, Shumway SD, Mizuarai S, Hirai H, Maitra A, Hidalgo M (2011) MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin Cancer Res 17(9):2799–2806. https://doi.org/10.1158/1078-0432.ccr-10-2580
Do K, Doroshow JH, Kummar S (2013) Wee1 kinase as a target for cancer therapy. Cell Cycle 12(19):3159–3164. https://doi.org/10.4161/cc.26062
Hassan M, Bielawski JP, Hempel JC, Waldman M (1996) Optimization and visualization of molecular diversity of combinatorial libraries. Mol Divers 2(1–2):64–74
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1):3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623. https://doi.org/10.1021/jm020017n
Squire CJ, Dickson JM, Ivanovic I, Baker EN (2005) Structure and inhibition of the human cell cycle checkpoint kinase, Wee1A kinase: an atypical tyrosine kinase with a key role in CDK1 regulation. Structure 13(4):541–550. https://doi.org/10.1016/j.str.2004.12.017
Smaill JB, Baker EN, Booth RJ, Bridges AJ, Dickson JM, Dobrusin EM, Ivanovic I, Kraker AJ, Lee HH, Lunney EA, Ortwine DF, Palmer BD, Quin J 3rd, Squire CJ, Thompson AM, Denny WA (2008) Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases. Eur J Med Chem 43(6):1276–1296. https://doi.org/10.1016/j.ejmech.2007.07.016
Smaill JB, Lee HH, Palmer BD, Thompson AM, Squire CJ, Baker EN, Booth RJ, Kraker A, Hook K, Denny WA (2008) Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases. Bioorg Med Chem Lett 18(3):929–933. https://doi.org/10.1016/j.bmcl.2007.12.046
Yang SY (2010) Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today 15(11–12):444–450. https://doi.org/10.1016/j.drudis.2010.03.013
Barnum D, Greene J, Smellie A, Sprague P (1996) Identification of common functional configurations among molecules. J Chem Inf Comput Sci 36(3):563–571. https://doi.org/10.1021/ci950273r
Thangapandian S, John S, Sakkiah S, Lee KW (2011) Potential virtual lead identification in the discovery of renin inhibitors: application of ligand and structure-based pharmacophore modeling approaches. Eur J Med Chem 46(6):2469–2476. https://doi.org/10.1016/j.ejmech.2011.03.035
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55(14):6582–6594. https://doi.org/10.1021/jm300687e
Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53(7):2719–2740. https://doi.org/10.1021/jm901137j
Lagorce D, Maupetit J, Baell J, Sperandio O, Tuffery P, Miteva MA, Galons H, Villoutreix BO (2011) The FAF-Drugs2 server: a multistep engine to prepare electronic chemical compound collections. Bioinformatics 27(14):2018–2020. https://doi.org/10.1093/bioinformatics/btr333
Jones G, Willett P, Glen RC, Leach AR, Taylor R by Cohen FE (1997) Development and validation of a genetic algorithm for flexible docking 11 edited. J Mol Biol 267(3):727–748. https://doi.org/10.1006/jmbi.1996.0897
Jones G, Willett P, Glen RC (1995) A genetic algorithm for flexible molecular overlay and pharmacophore elucidation. J Comput Aided Mol Des 9(6):532–549. https://doi.org/10.1007/BF00124324
Wichapong K, Lindner M, Pianwanit S, Kokpol S, Sippl W (2009) Receptor-based 3D-QSAR studies of checkpoint Wee1 kinase inhibitors. Eur J Med Chem 44(4):1383–1395. https://doi.org/10.1016/j.ejmech.2008.09.027
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37(24):4130–4146. https://doi.org/10.1021/jm00050a010
Manne R (1987) Analysis of two partial-least-squares algorithms for multivariate calibration. Chemometr Intell Lab Syst 2(1):187–197. https://doi.org/10.1016/0169-7439(87)80096-5
Lineweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56(3):658–666. https://doi.org/10.1021/ja01318a036
Cory AH, Owen TC, Barltrop JA, Cory JG (1991) Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun 3(7):207–212. https://doi.org/10.3727/095535491820873191
Barltrop JA, Owen TC, Cory AH, Cory JG (1991) 5-(3-carboxymethoxyphenyl)-2-(4,5-dimethylthiazolyl)-3-(4-sulfophenyl)tetrazolium, inner salt (MTS) and related analogs of 3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT) reducing to purple water-soluble formazans As cell-viability indicato. Bioorg Med Chem Lett 1(11):611–614
Riss TL, Moravec RA (1992) Comparison of MTT, XTT, and a novel tetrazolium compound MTS for in vitro proliferation and chemosensitivity assays. Mol Biol Cell 3(1):184–190
Mir SE, De Witt Hamer PC, Krawczyk PM, Balaj L, Claes A, Niers JM, Van Tilborg AA, Zwinderman AH, Geerts D, Kaspers GJ, Peter Vandertop W, Cloos J, Tannous BA, Wesseling P, Aten JA, Noske DP, Van Noorden CJ, Wurdinger T (2010) In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer cell 18(3):244–257. https://doi.org/10.1016/j.ccr.2010.08.011
PosthumaDeBoer J, Wurdinger T, Graat HC, van Beusechem VW, Helder MN, van Royen BJ, Kaspers GJ (2011) WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer 11:156. https://doi.org/10.1186/1471-2407-11-156
Yoshida T (2004) The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol 15(2):252–256. https://doi.org/10.1093/annonc/mdh073
Masaki T, Shiratori Y, Rengifo W, Igarashi K, Yamagata M, Kurokohchi K, Uchida N, Miyauchi Y, Yoshiji H, Watanabe S, Omata M, Kuriyama S (2003) Cyclins and cyclin-dependent kinases: comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology 37(3):534–543. https://doi.org/10.1053/jhep.2003.50112
Lin CC, Lin SY, Chung JG, Lin JP, Chen GW, Kao ST (2006) Down-regulation of cyclin B1 and up-regulation of Wee1 by berberine promotes entry of leukemia cells into the G2/M-phase of the cell cycle. Anticancer Res 26(2A):1097–1104
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. https://doi.org/10.1002/jcc.20291
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65(3):712–725. https://doi.org/10.1002/prot.21123
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174. https://doi.org/10.1002/jcc.20035
Wagner AB (2006) SciFinder Scholar 2006: an empirical analysis of research topic query processing. J Chem Inf Model 46(2):767–774. https://doi.org/10.1021/ci050481b
Acknowledgements
This work was financially supported by CAS “Light of West China” Program ([2014]91 to Z.Z.), CAS Strategic biological resources service network (ZSTH-021), the Strategic Priority Research Program of the Chinese Academy of Sciences, Grant No. XDA12030206.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Pu, Y., Liu, H. et al. Discovery of novel wee1 inhibitors via structure-based virtual screening and biological evaluation. J Comput Aided Mol Des 32, 901–915 (2018). https://doi.org/10.1007/s10822-018-0122-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0122-1