Abstract
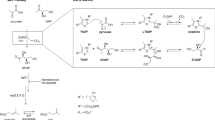
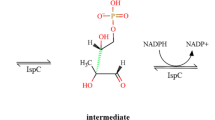
Proteins of the independent mevalonate pathway for isoprenoid biosynthesis are important targets for the development of new antibacterial compounds as this pathway is present in most pathogenic organisms such as Mycobacterium tuberculosis, dPlasmodium falciparum and Escherichia coli, but is not present in mammalian species, including humans. Deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) is an important target in this pathway and the most effective DXR inhibitor to date is fosmidomycin, which is used to treat malaria and, more recently, tuberculosis. Recently, Armstrong C. M. et al. showed that a mutant of DXR, S222T, induces a loss of the fosmidomycin inhibition efficiency, even though the bacteria culture is still viable and able to produce isoprenoids. As this represents a potential fosmidomycin-resistant mutation, it is important to understand the mechanism of this apparent mutation-induced resistance to fosmidomycin. Here, we used molecular dynamics simulations and Molecular Mechanics/Poisson Boltzmann Surface Area analysis to understand the structural and energetic basis of the resistance. Our results suggest that the point mutation results in changes to the structural dynamics of an active site loop that probably protects the active site and facilitates enzymatic reaction. From the simulation analysis, we also showed that the mutation results in changes in the interaction energy profiles in a way that can explain the observed activity of the mutant protein toward the natural inhibitor deoxy-d-xylulose 5-phosphate. These results should be taken into consideration in future efforts to develop new therapeutic antibiotic compounds that target DXR.







Similar content being viewed by others
References
Blair JM et al (2015) Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13(1):42–51
Kuntz L et al (2005) Isoprenoid biosynthesis as a target for antibacterial and antiparasitic drugs: phosphonohydroxamic acids as inhibitors of deoxyxylulose phosphate reducto-isomerase. Biochem J 386:127–135
Zhu W et al (2013) Antibacterial drug leads targeting isoprenoid biosynthesis. Proc Natl Acad Sci USA 110(1):123–128
Zhao LS et al (2013) Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem 82:497–530
Rohmer M (1999) The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep 16(5):565–574
Rohmer M et al (1993) Isoprenoid biosynthesis in bacteria—a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295:517–524
Rohmer M et al (1996) Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J Am Chem Soc 118(11):2564–2566
Rodriguez-Concepion M (2004) The MEP pathway: A new target for the development of herbicides, antibiotics and antimalarial drugs. Curr Pharm Des 10(19):2391–2400
Oldfield E (2010) Targeting isoprenoid biosynthesis for drug Discovery: bench to bedside. Acc Chem Res 43(9):1216–1226
Argyrou A, Blanchard JS (2004) Kinetic and chemical mechanism of mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate isomeroreductase. Biochemistry 43(14):4375–4384
Berman HM et al (2000) The protein data bank. Nucleic Acids Res 28(1):235–242
Mac Sweeney A, et al. (2005) The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J Mol Biol 345(1), 115–127.
Henriksson LM et al (2007) Structures of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate reductoisomerase provide new insights into catalysis. J Biol Chem 282(27):19905–19916
Williams SL, Andrew McCammon J (2009) Conformational dynamics of the flexible catalytic loop in Mycobacterium tuberculosis 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Chem Biol Drug Des 73(1):26–38
Manhas A et al (2019) Identification of natural compound inhibitors against PfDXR: a hybrid structure-based molecular modeling approach and molecular dynamics simulation studies. J Cell Biochem 120(9):14531–14543
Kuzuyama T et al (1998) Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett 39(43):7913–7916
Okuhara M, et al. (1980) Studies on new phosphonic acid antibiotics.1. Fr-900098, isolation and characterization. J Antibiot 33(1), 13–17.
Koppisch AT et al (2002) E-coli MEP synthase: steady-state kinetic analysis and substrate binding. Biochemistry 41(1):236–243
Zingle C et al (2010) Isoprenoid biosynthesis via the methylerythritol phosphate pathway: structural variations around phosphonate anchor and spacer of fosmidomycin, a potent inhibitor of deoxyxylulose phosphate reductoisomerase. J Org Chem 75(10):3203–3207
Jackson ER, Dowd CS (2012) Inhibition of 1-Deoxy-d-Xylulose-5-Phosphate reductoisomerase (Dxr): a review of the synthesis and biological evaluation of recent inhibitors. Curr Top Med Chem 12(7):706–728
Kuemmerle HP, Murakawa T, De Santis F (1987) Pharmacokinetic evaluation of fosmidomycin, a new phosphonic acid antibiotic. Chemioterapia 6(2):113–119
Brown AC, Parish T (2008) Dxr is essential in Mycobacterium tuberculosis and fosmidomycin resistance is due to a lack of uptake. BMC Microbiol 8(1):78
Kojo H, Shigi Y, Nishida M (1980) FR-31564, a new phosphonic acid antibiotic: bacterial resistance and membrane permeability. J Antibiot (Tokyo) 33(1):44–48
Martinez JL, Baquero F (2000) Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44(7):1771–1777
Armstrong CM et al (2015) Resistance to the antimicrobial agent fosmidomycin and an FR900098 prodrug through mutations in the deoxyxylulose phosphate reductoisomerase gene (dxr). Antimicrob Agents Chemother 59(9):5511–5519
Kollman PA et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33(12):889–897
Lafont V et al (2007) Protein–protein recognition and interaction hot spots in an antigen-antibody complex: free energy decomposition identifies "efficient amino acids". Proteins 67(2):418–434
Perez-Gil J et al (2012) Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis. J Biol Chem 287(19):15803–15809
Gagne D et al (2015) Perturbation of the conformational dynamics of an active-site loop alters enzyme activity. Structure 23(12):2256–2266
Wang Y, Berlow RB, Loria JP (2009) Role of loop-loop interactions in coordinating motions and enzymatic function in triosephosphate isomerase. Biochemistry 48(21):4548–4556
Zhang Z, et al. (1994) Crystal structure of recombinant chicken triosephosphate isomerase-phosphoglycolohydroxamate complex at 1.8-A resolution. Biochemistry 33(10), 2830–2837.
Browning C et al (2007) Critical role of desolvation in the binding of 20-hydroxyecdysone to the ecdysone receptor. J Biol Chem 282(45):32924–32934
Moroy G et al (2009) Molecular basis for Bcl-2 homology 3 domain recognition in the Bcl-2 protein family: identification of conserved hot spot interactions. J Biol Chem 284(26):17499–17511
Zanier K et al (2013) Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science 339(6120):694–698
Wilcoxon F (1945) Individual Comparisons by Ranking Methods. Biom Bull 1(6):80–83
Chen VB et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Pt 1):12–21
Eswar N et al (2006) Comparative protein structure modeling using modeller. Curr Protoc Bioinform 15(1):5–6
Hanwell MD et al (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4(1):17
Morris GM et al (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Chen D et al (2007) Accounting for ligand-bound metal ions in docking small molecules on adenylyl cyclase toxins. Proteins 67(3):593–605
MacKerell ADJ et al (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem 102:3586–3616
Vanommeslaeghe K et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690
Vanommeslaeghe, K. and A.D.J. MacKerell. Available from: https://cgenff.paramchem.org.
Mayne CG et al (2013) Rapid parameterization of small molecules using the force field toolkit. J Comput Chem 34(32):2757–2770
Frisch, M.J., et al., Gaussian 09 Rev. A.02. 2016: Wallingford, CT.
Brooks BR et al (2009) CHARMM: the biomolecular simulation program. J Comput Chem 26:1781–1802
Phillips JC et al (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Basconi JE, Shirts MR (2013) Effects of temperature control algorithms on transport properties and kinetics in molecular dynamics simulations. J Chem Theory Comput 9(7):2887–2899
Allen MP, Tildesley DJ (1987) Computer simulation of liquids. Oxford University Press, Oxford
Genheden S, Ryde U (2010) How to Obtain Statistically Converged MM/GBSA Results. J Comput Chem 31(4):837–846
Radykewicz T et al (2000) Biosynthesis of terpenoids: 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Escherichia coli is a class B dehydrogenase. FEBS Lett 465(2–3):157–160
Fox DT, Poulter CD (2005) Mechanistic studies with 2-C-methyl-D-erythritol 4-phosphate synthase from Escherichia coli. Biochemistry 44(23):8360–8368
Acknowledgements
This work was supported by funds from the Centre National de Recherche Scientifique (CNRS), Institut National de La Santé et de La Recherche Médicale (INSERM), Université de Strasbourg, the Agence Nationale de la Recherche (ANR), and by the Ministère de l’Enseignement Supérieur de la Recherche et de l’Innovation. Computing resources were provided by the Institut du Développement et des Ressources en Informatique Scientifique (IDRIS), the Centre Informatique National de l’Enseignement Supérieur (CINES) and the Méso-centre de Calcul de l’Université de Strasbourg supported by the national Equipex project EQUIP@ MESO. The authors thank L. Bianchetti for his help in the statistical analysis of the MM/PBSA results.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krebs, F.S., Esque, J. & Stote, R.H. A computational study of the molecular basis of antibiotic resistance in a DXR mutant. J Comput Aided Mol Des 33, 927–940 (2019). https://doi.org/10.1007/s10822-019-00229-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00229-5